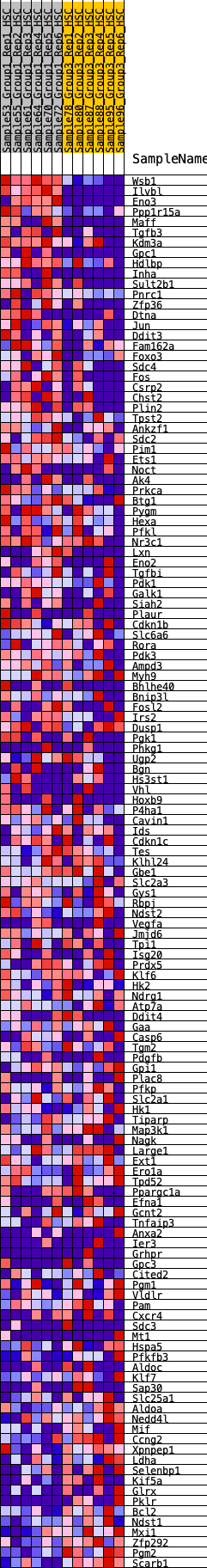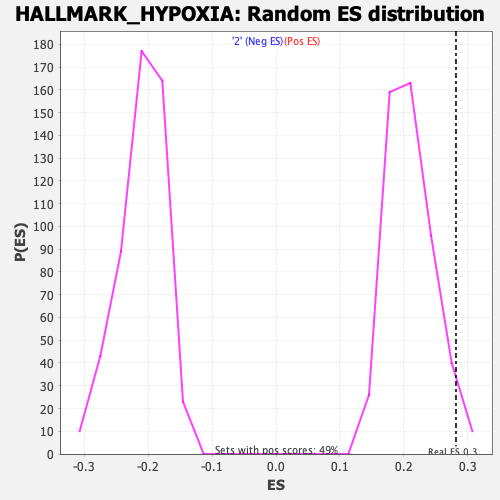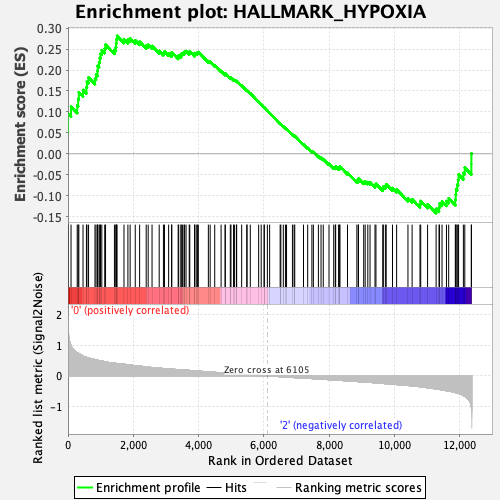
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group3.HSC_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_HYPOXIA |
| Enrichment Score (ES) | 0.2814011 |
| Normalized Enrichment Score (NES) | 1.3456987 |
| Nominal p-value | 0.04453441 |
| FDR q-value | 0.7760559 |
| FWER p-Value | 0.727 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Wsb1 | 0 | 2.165 | 0.0522 | Yes |
| 2 | Ilvbl | 2 | 1.823 | 0.0960 | Yes |
| 3 | Eno3 | 93 | 0.993 | 0.1126 | Yes |
| 4 | Ppp1r15a | 282 | 0.760 | 0.1155 | Yes |
| 5 | Maff | 311 | 0.746 | 0.1312 | Yes |
| 6 | Tgfb3 | 332 | 0.729 | 0.1471 | Yes |
| 7 | Kdm3a | 460 | 0.650 | 0.1524 | Yes |
| 8 | Gpc1 | 564 | 0.608 | 0.1586 | Yes |
| 9 | Hdlbp | 580 | 0.603 | 0.1719 | Yes |
| 10 | Inha | 629 | 0.584 | 0.1821 | Yes |
| 11 | Sult2b1 | 826 | 0.536 | 0.1790 | Yes |
| 12 | Pnrc1 | 859 | 0.526 | 0.1890 | Yes |
| 13 | Zfp36 | 904 | 0.514 | 0.1978 | Yes |
| 14 | Dtna | 909 | 0.513 | 0.2098 | Yes |
| 15 | Jun | 952 | 0.502 | 0.2185 | Yes |
| 16 | Ddit3 | 975 | 0.497 | 0.2287 | Yes |
| 17 | Fam162a | 995 | 0.493 | 0.2390 | Yes |
| 18 | Foxo3 | 1035 | 0.482 | 0.2474 | Yes |
| 19 | Sdc4 | 1125 | 0.465 | 0.2513 | Yes |
| 20 | Fos | 1149 | 0.460 | 0.2606 | Yes |
| 21 | Csrp2 | 1422 | 0.414 | 0.2483 | Yes |
| 22 | Chst2 | 1465 | 0.410 | 0.2547 | Yes |
| 23 | Plin2 | 1474 | 0.409 | 0.2639 | Yes |
| 24 | Tpst2 | 1484 | 0.407 | 0.2730 | Yes |
| 25 | Ankzf1 | 1502 | 0.406 | 0.2814 | Yes |
| 26 | Sdc2 | 1714 | 0.380 | 0.2733 | No |
| 27 | Pim1 | 1834 | 0.365 | 0.2724 | No |
| 28 | Ets1 | 1902 | 0.355 | 0.2755 | No |
| 29 | Noct | 2059 | 0.333 | 0.2707 | No |
| 30 | Ak4 | 2193 | 0.318 | 0.2675 | No |
| 31 | Prkca | 2394 | 0.294 | 0.2583 | No |
| 32 | Btg1 | 2455 | 0.286 | 0.2603 | No |
| 33 | Pygm | 2573 | 0.275 | 0.2573 | No |
| 34 | Hexa | 2792 | 0.254 | 0.2456 | No |
| 35 | Pfkl | 2922 | 0.241 | 0.2409 | No |
| 36 | Nr3c1 | 2953 | 0.238 | 0.2442 | No |
| 37 | Lxn | 3081 | 0.227 | 0.2392 | No |
| 38 | Eno2 | 3173 | 0.221 | 0.2371 | No |
| 39 | Tgfbi | 3176 | 0.221 | 0.2423 | No |
| 40 | Pdk1 | 3373 | 0.205 | 0.2312 | No |
| 41 | Galk1 | 3389 | 0.204 | 0.2349 | No |
| 42 | Siah2 | 3458 | 0.201 | 0.2342 | No |
| 43 | Plaur | 3465 | 0.200 | 0.2385 | No |
| 44 | Cdkn1b | 3503 | 0.197 | 0.2402 | No |
| 45 | Slc6a6 | 3551 | 0.193 | 0.2410 | No |
| 46 | Rora | 3570 | 0.191 | 0.2442 | No |
| 47 | Pdk3 | 3614 | 0.188 | 0.2452 | No |
| 48 | Ampd3 | 3712 | 0.179 | 0.2416 | No |
| 49 | Myh9 | 3728 | 0.178 | 0.2446 | No |
| 50 | Bhlhe40 | 3877 | 0.165 | 0.2365 | No |
| 51 | Bnip3l | 3878 | 0.165 | 0.2405 | No |
| 52 | Fosl2 | 3926 | 0.161 | 0.2405 | No |
| 53 | Irs2 | 3962 | 0.158 | 0.2415 | No |
| 54 | Dusp1 | 3990 | 0.156 | 0.2430 | No |
| 55 | Pgk1 | 4297 | 0.131 | 0.2211 | No |
| 56 | Phkg1 | 4352 | 0.127 | 0.2198 | No |
| 57 | Ugp2 | 4492 | 0.116 | 0.2112 | No |
| 58 | Bgn | 4687 | 0.099 | 0.1977 | No |
| 59 | Hs3st1 | 4812 | 0.091 | 0.1898 | No |
| 60 | Vhl | 4813 | 0.091 | 0.1920 | No |
| 61 | Hoxb9 | 4969 | 0.079 | 0.1812 | No |
| 62 | P4ha1 | 4989 | 0.078 | 0.1815 | No |
| 63 | Cavin1 | 5068 | 0.071 | 0.1769 | No |
| 64 | Ids | 5100 | 0.069 | 0.1760 | No |
| 65 | Cdkn1c | 5157 | 0.065 | 0.1730 | No |
| 66 | Tes | 5164 | 0.064 | 0.1740 | No |
| 67 | Klhl24 | 5317 | 0.054 | 0.1629 | No |
| 68 | Gbe1 | 5474 | 0.043 | 0.1512 | No |
| 69 | Slc2a3 | 5481 | 0.042 | 0.1517 | No |
| 70 | Gys1 | 5581 | 0.036 | 0.1445 | No |
| 71 | Rbpj | 5837 | 0.017 | 0.1240 | No |
| 72 | Ndst2 | 5915 | 0.012 | 0.1180 | No |
| 73 | Vegfa | 6004 | 0.006 | 0.1110 | No |
| 74 | Jmjd6 | 6009 | 0.006 | 0.1108 | No |
| 75 | Tpi1 | 6104 | 0.000 | 0.1031 | No |
| 76 | Isg20 | 6173 | -0.002 | 0.0976 | No |
| 77 | Prdx5 | 6495 | -0.025 | 0.0719 | No |
| 78 | Klf6 | 6516 | -0.026 | 0.0709 | No |
| 79 | Hk2 | 6592 | -0.031 | 0.0655 | No |
| 80 | Ndrg1 | 6657 | -0.036 | 0.0612 | No |
| 81 | Atp7a | 6691 | -0.039 | 0.0594 | No |
| 82 | Ddit4 | 6871 | -0.050 | 0.0460 | No |
| 83 | Gaa | 6921 | -0.054 | 0.0433 | No |
| 84 | Casp6 | 6942 | -0.055 | 0.0430 | No |
| 85 | Tgm2 | 7210 | -0.071 | 0.0229 | No |
| 86 | Pdgfb | 7338 | -0.080 | 0.0144 | No |
| 87 | Gpi1 | 7467 | -0.088 | 0.0060 | No |
| 88 | Plac8 | 7512 | -0.091 | 0.0046 | No |
| 89 | Pfkp | 7666 | -0.102 | -0.0054 | No |
| 90 | Slc2a1 | 7747 | -0.108 | -0.0094 | No |
| 91 | Hk1 | 7815 | -0.112 | -0.0121 | No |
| 92 | Tiparp | 7991 | -0.123 | -0.0235 | No |
| 93 | Map3k1 | 8135 | -0.133 | -0.0320 | No |
| 94 | Nagk | 8183 | -0.136 | -0.0325 | No |
| 95 | Large1 | 8195 | -0.137 | -0.0301 | No |
| 96 | Ext1 | 8284 | -0.144 | -0.0338 | No |
| 97 | Ero1a | 8320 | -0.146 | -0.0332 | No |
| 98 | Tpd52 | 8321 | -0.146 | -0.0297 | No |
| 99 | Ppargc1a | 8557 | -0.161 | -0.0450 | No |
| 100 | Efna1 | 8844 | -0.184 | -0.0640 | No |
| 101 | Gcnt2 | 8888 | -0.187 | -0.0630 | No |
| 102 | Tnfaip3 | 8893 | -0.188 | -0.0588 | No |
| 103 | Anxa2 | 9053 | -0.199 | -0.0670 | No |
| 104 | Ier3 | 9098 | -0.203 | -0.0657 | No |
| 105 | Grhpr | 9179 | -0.209 | -0.0672 | No |
| 106 | Gpc3 | 9247 | -0.213 | -0.0676 | No |
| 107 | Cited2 | 9399 | -0.227 | -0.0744 | No |
| 108 | Pgm1 | 9428 | -0.229 | -0.0712 | No |
| 109 | Vldlr | 9637 | -0.247 | -0.0823 | No |
| 110 | Pam | 9654 | -0.248 | -0.0776 | No |
| 111 | Cxcr4 | 9717 | -0.253 | -0.0766 | No |
| 112 | Sdc3 | 9741 | -0.255 | -0.0723 | No |
| 113 | Mt1 | 9935 | -0.272 | -0.0815 | No |
| 114 | Hspa5 | 10061 | -0.284 | -0.0849 | No |
| 115 | Pfkfb3 | 10408 | -0.314 | -0.1056 | No |
| 116 | Aldoc | 10535 | -0.328 | -0.1080 | No |
| 117 | Klf7 | 10776 | -0.355 | -0.1191 | No |
| 118 | Sap30 | 10799 | -0.358 | -0.1123 | No |
| 119 | Slc25a1 | 11009 | -0.389 | -0.1200 | No |
| 120 | Aldoa | 11270 | -0.429 | -0.1309 | No |
| 121 | Nedd4l | 11359 | -0.442 | -0.1275 | No |
| 122 | Mif | 11375 | -0.445 | -0.1180 | No |
| 123 | Ccng2 | 11451 | -0.460 | -0.1130 | No |
| 124 | Xpnpep1 | 11587 | -0.485 | -0.1124 | No |
| 125 | Ldha | 11655 | -0.498 | -0.1058 | No |
| 126 | Selenbp1 | 11858 | -0.547 | -0.1092 | No |
| 127 | Kif5a | 11871 | -0.549 | -0.0969 | No |
| 128 | Glrx | 11880 | -0.552 | -0.0843 | No |
| 129 | Pklr | 11915 | -0.564 | -0.0735 | No |
| 130 | Bcl2 | 11943 | -0.574 | -0.0619 | No |
| 131 | Ndst1 | 11958 | -0.577 | -0.0491 | No |
| 132 | Mxi1 | 12105 | -0.651 | -0.0453 | No |
| 133 | Zfp292 | 12148 | -0.669 | -0.0327 | No |
| 134 | Pgm2 | 12349 | -1.038 | -0.0240 | No |
| 135 | Scarb1 | 12350 | -1.041 | 0.0011 | No |