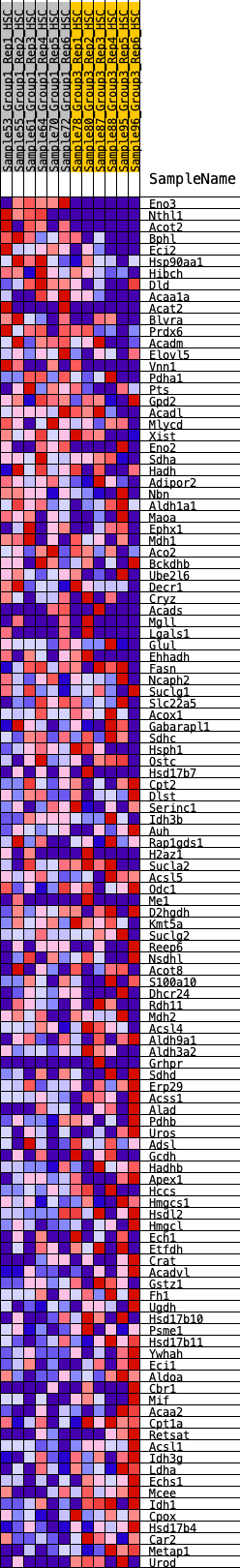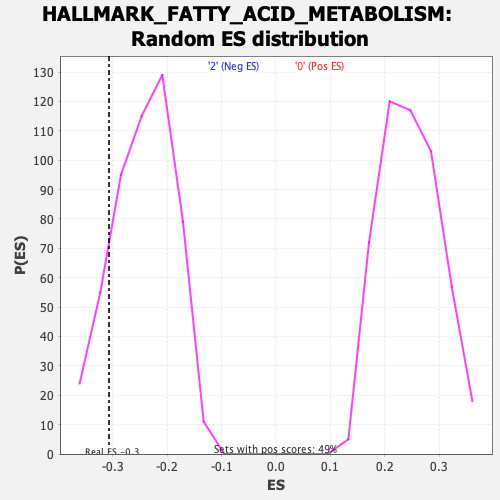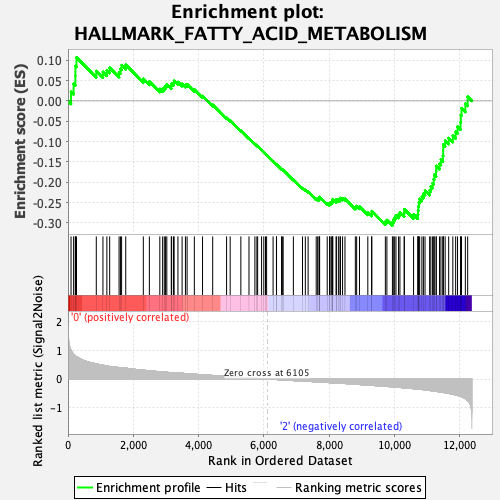
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group3.HSC_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
| Enrichment Score (ES) | -0.30656984 |
| Normalized Enrichment Score (NES) | -1.2556415 |
| Nominal p-value | 0.1515748 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.891 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Eno3 | 93 | 0.993 | 0.0225 | No |
| 2 | Nthl1 | 172 | 0.860 | 0.0423 | No |
| 3 | Acot2 | 225 | 0.796 | 0.0622 | No |
| 4 | Bphl | 227 | 0.794 | 0.0862 | No |
| 5 | Eci2 | 260 | 0.773 | 0.1070 | No |
| 6 | Hsp90aa1 | 866 | 0.525 | 0.0735 | No |
| 7 | Hibch | 1070 | 0.476 | 0.0714 | No |
| 8 | Dld | 1190 | 0.452 | 0.0754 | No |
| 9 | Acaa1a | 1276 | 0.433 | 0.0816 | No |
| 10 | Acat2 | 1564 | 0.397 | 0.0702 | No |
| 11 | Blvra | 1606 | 0.393 | 0.0788 | No |
| 12 | Prdx6 | 1641 | 0.390 | 0.0878 | No |
| 13 | Acadm | 1768 | 0.375 | 0.0889 | No |
| 14 | Elovl5 | 2306 | 0.305 | 0.0543 | No |
| 15 | Vnn1 | 2491 | 0.281 | 0.0478 | No |
| 16 | Pdha1 | 2811 | 0.252 | 0.0294 | No |
| 17 | Pts | 2897 | 0.243 | 0.0299 | No |
| 18 | Gpd2 | 2948 | 0.239 | 0.0330 | No |
| 19 | Acadl | 2987 | 0.234 | 0.0370 | No |
| 20 | Mlycd | 3025 | 0.231 | 0.0410 | No |
| 21 | Xist | 3162 | 0.223 | 0.0367 | No |
| 22 | Eno2 | 3173 | 0.221 | 0.0426 | No |
| 23 | Sdha | 3231 | 0.217 | 0.0445 | No |
| 24 | Hadh | 3249 | 0.215 | 0.0496 | No |
| 25 | Adipor2 | 3366 | 0.206 | 0.0464 | No |
| 26 | Nbn | 3493 | 0.197 | 0.0421 | No |
| 27 | Aldh1a1 | 3595 | 0.189 | 0.0396 | No |
| 28 | Maoa | 3646 | 0.186 | 0.0412 | No |
| 29 | Ephx1 | 3868 | 0.165 | 0.0281 | No |
| 30 | Mdh1 | 4119 | 0.145 | 0.0121 | No |
| 31 | Aco2 | 4430 | 0.122 | -0.0095 | No |
| 32 | Bckdhb | 4855 | 0.087 | -0.0415 | No |
| 33 | Ube2l6 | 4965 | 0.079 | -0.0480 | No |
| 34 | Decr1 | 5292 | 0.055 | -0.0729 | No |
| 35 | Cryz | 5541 | 0.039 | -0.0920 | No |
| 36 | Acads | 5722 | 0.026 | -0.1059 | No |
| 37 | Mgll | 5783 | 0.022 | -0.1101 | No |
| 38 | Lgals1 | 5806 | 0.021 | -0.1113 | No |
| 39 | Glul | 5926 | 0.011 | -0.1207 | No |
| 40 | Ehhadh | 5994 | 0.007 | -0.1259 | No |
| 41 | Fasn | 6047 | 0.003 | -0.1301 | No |
| 42 | Ncaph2 | 6081 | 0.001 | -0.1328 | No |
| 43 | Suclg1 | 6279 | -0.009 | -0.1486 | No |
| 44 | Slc22a5 | 6382 | -0.016 | -0.1564 | No |
| 45 | Acox1 | 6384 | -0.017 | -0.1560 | No |
| 46 | Gabarapl1 | 6537 | -0.028 | -0.1676 | No |
| 47 | Sdhc | 6562 | -0.030 | -0.1686 | No |
| 48 | Hsph1 | 6587 | -0.031 | -0.1696 | No |
| 49 | Ostc | 6900 | -0.053 | -0.1935 | No |
| 50 | Hsd17b7 | 7180 | -0.070 | -0.2142 | No |
| 51 | Cpt2 | 7266 | -0.075 | -0.2189 | No |
| 52 | Dlst | 7351 | -0.080 | -0.2233 | No |
| 53 | Serinc1 | 7599 | -0.097 | -0.2405 | No |
| 54 | Idh3b | 7643 | -0.101 | -0.2409 | No |
| 55 | Auh | 7686 | -0.104 | -0.2412 | No |
| 56 | Rap1gds1 | 7697 | -0.104 | -0.2389 | No |
| 57 | H2az1 | 7704 | -0.104 | -0.2362 | No |
| 58 | Sucla2 | 7936 | -0.119 | -0.2515 | No |
| 59 | Acsl5 | 8007 | -0.123 | -0.2534 | No |
| 60 | Odc1 | 8020 | -0.124 | -0.2506 | No |
| 61 | Me1 | 8060 | -0.128 | -0.2500 | No |
| 62 | D2hgdh | 8086 | -0.130 | -0.2481 | No |
| 63 | Kmt5a | 8100 | -0.131 | -0.2452 | No |
| 64 | Suclg2 | 8106 | -0.131 | -0.2416 | No |
| 65 | Reep6 | 8210 | -0.139 | -0.2458 | No |
| 66 | Nsdhl | 8215 | -0.139 | -0.2419 | No |
| 67 | Acot8 | 8278 | -0.144 | -0.2426 | No |
| 68 | S100a10 | 8327 | -0.147 | -0.2421 | No |
| 69 | Dhcr24 | 8341 | -0.148 | -0.2387 | No |
| 70 | Rdh11 | 8406 | -0.152 | -0.2393 | No |
| 71 | Mdh2 | 8480 | -0.156 | -0.2405 | No |
| 72 | Acsl4 | 8797 | -0.180 | -0.2609 | No |
| 73 | Aldh9a1 | 8837 | -0.183 | -0.2585 | No |
| 74 | Aldh3a2 | 8924 | -0.190 | -0.2597 | No |
| 75 | Grhpr | 9179 | -0.209 | -0.2742 | No |
| 76 | Sdhd | 9293 | -0.217 | -0.2768 | No |
| 77 | Erp29 | 9303 | -0.217 | -0.2710 | No |
| 78 | Acss1 | 9716 | -0.253 | -0.2969 | No |
| 79 | Alad | 9763 | -0.258 | -0.2929 | No |
| 80 | Pdhb | 9932 | -0.272 | -0.2983 | Yes |
| 81 | Uros | 9959 | -0.274 | -0.2921 | Yes |
| 82 | Adsl | 10001 | -0.277 | -0.2871 | Yes |
| 83 | Gcdh | 10041 | -0.281 | -0.2817 | Yes |
| 84 | Hadhb | 10120 | -0.287 | -0.2794 | Yes |
| 85 | Apex1 | 10162 | -0.292 | -0.2739 | Yes |
| 86 | Hccs | 10293 | -0.304 | -0.2753 | Yes |
| 87 | Hmgcs1 | 10297 | -0.304 | -0.2663 | Yes |
| 88 | Hsdl2 | 10580 | -0.334 | -0.2792 | Yes |
| 89 | Hmgcl | 10710 | -0.348 | -0.2791 | Yes |
| 90 | Ech1 | 10718 | -0.348 | -0.2691 | Yes |
| 91 | Etfdh | 10728 | -0.349 | -0.2593 | Yes |
| 92 | Crat | 10742 | -0.351 | -0.2497 | Yes |
| 93 | Acadvl | 10767 | -0.354 | -0.2409 | Yes |
| 94 | Gstz1 | 10835 | -0.362 | -0.2354 | Yes |
| 95 | Fh1 | 10881 | -0.368 | -0.2279 | Yes |
| 96 | Ugdh | 10931 | -0.376 | -0.2205 | Yes |
| 97 | Hsd17b10 | 11072 | -0.397 | -0.2199 | Yes |
| 98 | Psme1 | 11108 | -0.403 | -0.2106 | Yes |
| 99 | Hsd17b11 | 11162 | -0.411 | -0.2024 | Yes |
| 100 | Ywhah | 11197 | -0.416 | -0.1926 | Yes |
| 101 | Eci1 | 11214 | -0.419 | -0.1812 | Yes |
| 102 | Aldoa | 11270 | -0.429 | -0.1727 | Yes |
| 103 | Cbr1 | 11274 | -0.429 | -0.1599 | Yes |
| 104 | Mif | 11375 | -0.445 | -0.1546 | Yes |
| 105 | Acaa2 | 11417 | -0.454 | -0.1441 | Yes |
| 106 | Cpt1a | 11479 | -0.465 | -0.1350 | Yes |
| 107 | Retsat | 11487 | -0.466 | -0.1214 | Yes |
| 108 | Acsl1 | 11488 | -0.466 | -0.1073 | Yes |
| 109 | Idh3g | 11547 | -0.478 | -0.0976 | Yes |
| 110 | Ldha | 11655 | -0.498 | -0.0912 | Yes |
| 111 | Echs1 | 11782 | -0.526 | -0.0855 | Yes |
| 112 | Mcee | 11868 | -0.549 | -0.0758 | Yes |
| 113 | Idh1 | 11926 | -0.566 | -0.0632 | Yes |
| 114 | Cpox | 12018 | -0.601 | -0.0525 | Yes |
| 115 | Hsd17b4 | 12028 | -0.604 | -0.0349 | Yes |
| 116 | Car2 | 12048 | -0.612 | -0.0178 | Yes |
| 117 | Metap1 | 12165 | -0.681 | -0.0066 | Yes |
| 118 | Urod | 12236 | -0.749 | 0.0104 | Yes |