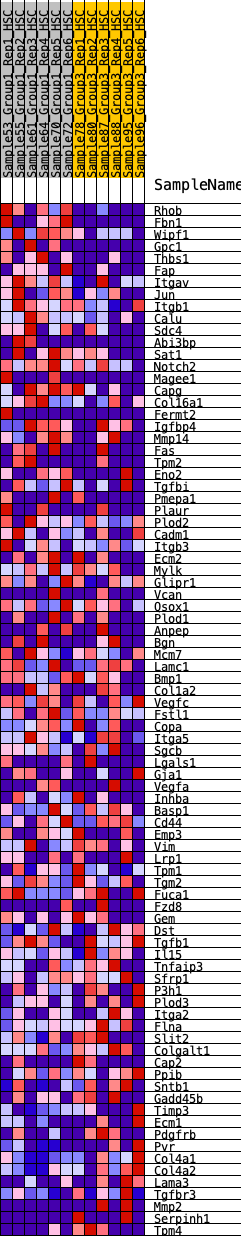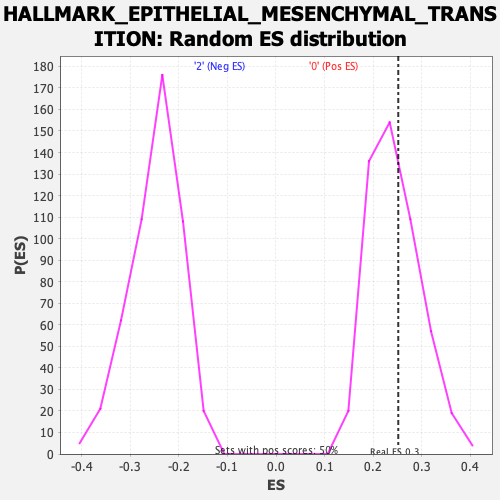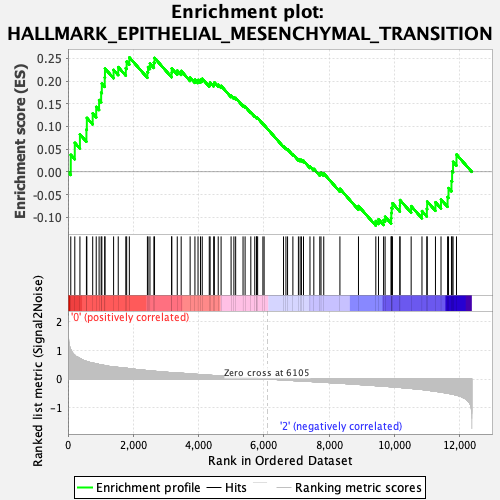
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group3.HSC_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | 0.2520757 |
| Normalized Enrichment Score (NES) | 1.0305713 |
| Nominal p-value | 0.40480962 |
| FDR q-value | 0.861015 |
| FWER p-Value | 0.997 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rhob | 85 | 1.009 | 0.0375 | Yes |
| 2 | Fbn1 | 208 | 0.825 | 0.0640 | Yes |
| 3 | Wipf1 | 365 | 0.709 | 0.0825 | Yes |
| 4 | Gpc1 | 564 | 0.608 | 0.0932 | Yes |
| 5 | Thbs1 | 577 | 0.605 | 0.1189 | Yes |
| 6 | Fap | 756 | 0.553 | 0.1287 | Yes |
| 7 | Itgav | 864 | 0.525 | 0.1432 | Yes |
| 8 | Jun | 952 | 0.502 | 0.1582 | Yes |
| 9 | Itgb1 | 1015 | 0.490 | 0.1747 | Yes |
| 10 | Calu | 1036 | 0.482 | 0.1943 | Yes |
| 11 | Sdc4 | 1125 | 0.465 | 0.2077 | Yes |
| 12 | Abi3bp | 1132 | 0.463 | 0.2276 | Yes |
| 13 | Sat1 | 1396 | 0.417 | 0.2246 | Yes |
| 14 | Notch2 | 1538 | 0.401 | 0.2307 | Yes |
| 15 | Magee1 | 1770 | 0.375 | 0.2284 | Yes |
| 16 | Capg | 1796 | 0.372 | 0.2428 | Yes |
| 17 | Col16a1 | 1878 | 0.360 | 0.2521 | Yes |
| 18 | Fermt2 | 2431 | 0.289 | 0.2199 | No |
| 19 | Igfbp4 | 2451 | 0.287 | 0.2310 | No |
| 20 | Mmp14 | 2509 | 0.279 | 0.2386 | No |
| 21 | Fas | 2630 | 0.269 | 0.2407 | No |
| 22 | Tpm2 | 2649 | 0.267 | 0.2510 | No |
| 23 | Eno2 | 3173 | 0.221 | 0.2181 | No |
| 24 | Tgfbi | 3176 | 0.221 | 0.2277 | No |
| 25 | Pmepa1 | 3345 | 0.208 | 0.2232 | No |
| 26 | Plaur | 3465 | 0.200 | 0.2223 | No |
| 27 | Plod2 | 3739 | 0.177 | 0.2079 | No |
| 28 | Cadm1 | 3886 | 0.164 | 0.2032 | No |
| 29 | Itgb3 | 3981 | 0.157 | 0.2025 | No |
| 30 | Ecm2 | 4057 | 0.150 | 0.2030 | No |
| 31 | Mylk | 4110 | 0.146 | 0.2052 | No |
| 32 | Glipr1 | 4325 | 0.129 | 0.1934 | No |
| 33 | Vcan | 4351 | 0.127 | 0.1970 | No |
| 34 | Qsox1 | 4460 | 0.119 | 0.1935 | No |
| 35 | Plod1 | 4481 | 0.117 | 0.1970 | No |
| 36 | Anpep | 4599 | 0.107 | 0.1922 | No |
| 37 | Bgn | 4687 | 0.099 | 0.1894 | No |
| 38 | Mcm7 | 4993 | 0.078 | 0.1680 | No |
| 39 | Lamc1 | 5079 | 0.070 | 0.1642 | No |
| 40 | Bmp1 | 5133 | 0.066 | 0.1628 | No |
| 41 | Col1a2 | 5359 | 0.051 | 0.1467 | No |
| 42 | Vegfc | 5422 | 0.047 | 0.1437 | No |
| 43 | Fstl1 | 5597 | 0.034 | 0.1311 | No |
| 44 | Copa | 5718 | 0.027 | 0.1225 | No |
| 45 | Itga5 | 5772 | 0.023 | 0.1191 | No |
| 46 | Sgcb | 5779 | 0.022 | 0.1196 | No |
| 47 | Lgals1 | 5806 | 0.021 | 0.1184 | No |
| 48 | Gja1 | 5967 | 0.009 | 0.1058 | No |
| 49 | Vegfa | 6004 | 0.006 | 0.1031 | No |
| 50 | Inhba | 6598 | -0.032 | 0.0562 | No |
| 51 | Basp1 | 6672 | -0.037 | 0.0519 | No |
| 52 | Cd44 | 6722 | -0.040 | 0.0497 | No |
| 53 | Emp3 | 6885 | -0.052 | 0.0388 | No |
| 54 | Vim | 7054 | -0.062 | 0.0278 | No |
| 55 | Lrp1 | 7107 | -0.066 | 0.0265 | No |
| 56 | Tpm1 | 7147 | -0.068 | 0.0263 | No |
| 57 | Tgm2 | 7210 | -0.071 | 0.0244 | No |
| 58 | Fuca1 | 7408 | -0.083 | 0.0120 | No |
| 59 | Fzd8 | 7526 | -0.092 | 0.0065 | No |
| 60 | Gem | 7709 | -0.105 | -0.0036 | No |
| 61 | Dst | 7746 | -0.108 | -0.0018 | No |
| 62 | Tgfb1 | 7830 | -0.113 | -0.0036 | No |
| 63 | Il15 | 8325 | -0.147 | -0.0374 | No |
| 64 | Tnfaip3 | 8893 | -0.188 | -0.0753 | No |
| 65 | Sfrp1 | 9424 | -0.229 | -0.1084 | No |
| 66 | P3h1 | 9506 | -0.236 | -0.1046 | No |
| 67 | Plod3 | 9662 | -0.249 | -0.1062 | No |
| 68 | Itga2 | 9708 | -0.253 | -0.0987 | No |
| 69 | Flna | 9893 | -0.268 | -0.1019 | No |
| 70 | Slit2 | 9898 | -0.268 | -0.0904 | No |
| 71 | Colgalt1 | 9910 | -0.270 | -0.0795 | No |
| 72 | Cap2 | 9933 | -0.272 | -0.0693 | No |
| 73 | Ppib | 10161 | -0.292 | -0.0749 | No |
| 74 | Sntb1 | 10164 | -0.292 | -0.0622 | No |
| 75 | Gadd45b | 10506 | -0.326 | -0.0756 | No |
| 76 | Timp3 | 10837 | -0.362 | -0.0865 | No |
| 77 | Ecm1 | 10986 | -0.386 | -0.0815 | No |
| 78 | Pdgfrb | 10997 | -0.387 | -0.0653 | No |
| 79 | Pvr | 11250 | -0.425 | -0.0671 | No |
| 80 | Col4a1 | 11420 | -0.455 | -0.0608 | No |
| 81 | Col4a2 | 11621 | -0.492 | -0.0554 | No |
| 82 | Lama3 | 11650 | -0.497 | -0.0358 | No |
| 83 | Tgfbr3 | 11740 | -0.517 | -0.0203 | No |
| 84 | Mmp2 | 11760 | -0.521 | 0.0012 | No |
| 85 | Serpinh1 | 11789 | -0.528 | 0.0222 | No |
| 86 | Tpm4 | 11894 | -0.556 | 0.0382 | No |