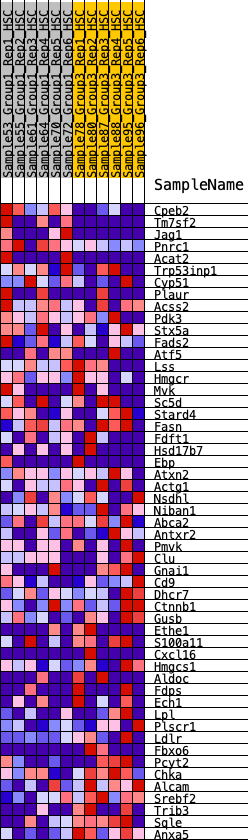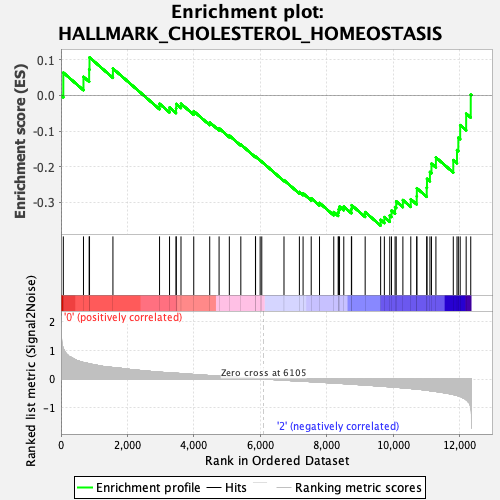
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group3.HSC_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | -0.36483952 |
| Normalized Enrichment Score (NES) | -1.2230452 |
| Nominal p-value | 0.1783567 |
| FDR q-value | 0.9890943 |
| FWER p-Value | 0.936 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cpeb2 | 70 | 1.058 | 0.0635 | No |
| 2 | Tm7sf2 | 677 | 0.572 | 0.0517 | No |
| 3 | Jag1 | 848 | 0.529 | 0.0725 | No |
| 4 | Pnrc1 | 859 | 0.526 | 0.1060 | No |
| 5 | Acat2 | 1564 | 0.397 | 0.0749 | No |
| 6 | Trp53inp1 | 2968 | 0.236 | -0.0237 | No |
| 7 | Cyp51 | 3269 | 0.213 | -0.0341 | No |
| 8 | Plaur | 3465 | 0.200 | -0.0369 | No |
| 9 | Acss2 | 3471 | 0.199 | -0.0242 | No |
| 10 | Pdk3 | 3614 | 0.188 | -0.0235 | No |
| 11 | Stx5a | 3998 | 0.155 | -0.0445 | No |
| 12 | Fads2 | 4479 | 0.117 | -0.0758 | No |
| 13 | Atf5 | 4761 | 0.094 | -0.0924 | No |
| 14 | Lss | 5070 | 0.071 | -0.1128 | No |
| 15 | Hmgcr | 5416 | 0.047 | -0.1378 | No |
| 16 | Mvk | 5857 | 0.016 | -0.1725 | No |
| 17 | Sc5d | 5860 | 0.015 | -0.1716 | No |
| 18 | Stard4 | 5998 | 0.007 | -0.1823 | No |
| 19 | Fasn | 6047 | 0.003 | -0.1860 | No |
| 20 | Fdft1 | 6716 | -0.040 | -0.2377 | No |
| 21 | Hsd17b7 | 7180 | -0.070 | -0.2707 | No |
| 22 | Ebp | 7291 | -0.077 | -0.2746 | No |
| 23 | Atxn2 | 7536 | -0.092 | -0.2884 | No |
| 24 | Actg1 | 7786 | -0.111 | -0.3014 | No |
| 25 | Nsdhl | 8215 | -0.139 | -0.3271 | No |
| 26 | Niban1 | 8348 | -0.148 | -0.3281 | No |
| 27 | Abca2 | 8362 | -0.149 | -0.3194 | No |
| 28 | Antxr2 | 8389 | -0.151 | -0.3117 | No |
| 29 | Pmvk | 8517 | -0.159 | -0.3116 | No |
| 30 | Clu | 8746 | -0.176 | -0.3187 | No |
| 31 | Gnai1 | 8754 | -0.177 | -0.3077 | No |
| 32 | Cd9 | 9161 | -0.208 | -0.3271 | No |
| 33 | Dhcr7 | 9627 | -0.246 | -0.3487 | Yes |
| 34 | Ctnnb1 | 9739 | -0.255 | -0.3410 | Yes |
| 35 | Gusb | 9902 | -0.269 | -0.3366 | Yes |
| 36 | Ethe1 | 9954 | -0.274 | -0.3229 | Yes |
| 37 | S100a11 | 10062 | -0.284 | -0.3130 | Yes |
| 38 | Cxcl16 | 10096 | -0.286 | -0.2969 | Yes |
| 39 | Hmgcs1 | 10297 | -0.304 | -0.2933 | Yes |
| 40 | Aldoc | 10535 | -0.328 | -0.2911 | Yes |
| 41 | Fdps | 10714 | -0.348 | -0.2828 | Yes |
| 42 | Ech1 | 10718 | -0.348 | -0.2602 | Yes |
| 43 | Lpl | 11015 | -0.389 | -0.2588 | Yes |
| 44 | Plscr1 | 11023 | -0.390 | -0.2338 | Yes |
| 45 | Ldlr | 11112 | -0.403 | -0.2146 | Yes |
| 46 | Fbxo6 | 11158 | -0.411 | -0.1914 | Yes |
| 47 | Pcyt2 | 11292 | -0.431 | -0.1740 | Yes |
| 48 | Chka | 11815 | -0.534 | -0.1814 | Yes |
| 49 | Alcam | 11927 | -0.567 | -0.1534 | Yes |
| 50 | Srebf2 | 11967 | -0.580 | -0.1186 | Yes |
| 51 | Trib3 | 12023 | -0.604 | -0.0836 | Yes |
| 52 | Sqle | 12203 | -0.717 | -0.0512 | Yes |
| 53 | Anxa5 | 12341 | -0.980 | 0.0018 | Yes |