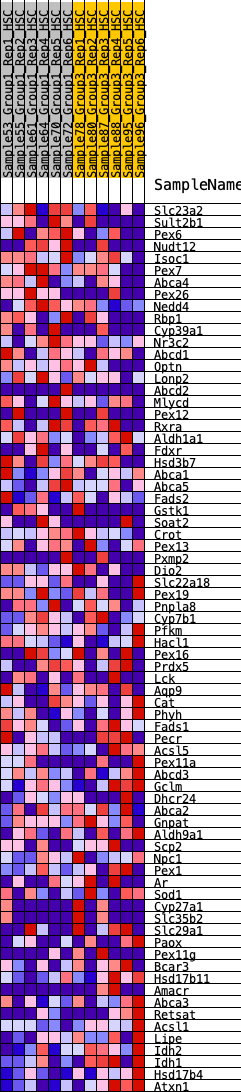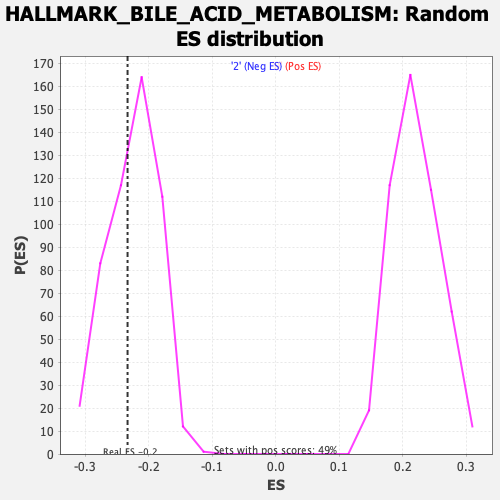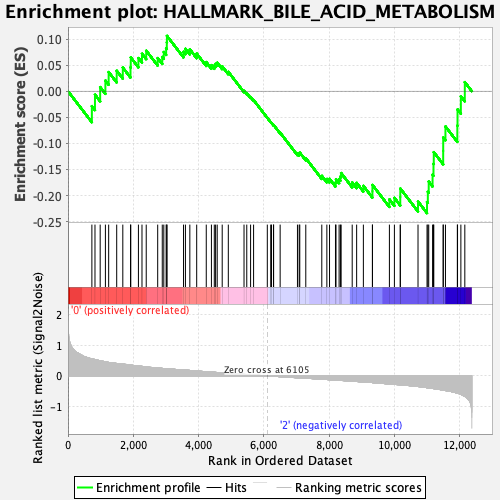
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group3.HSC_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.23366332 |
| Normalized Enrichment Score (NES) | -1.0366313 |
| Nominal p-value | 0.39411765 |
| FDR q-value | 0.6074643 |
| FWER p-Value | 0.996 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Slc23a2 | 731 | 0.559 | -0.0286 | No |
| 2 | Sult2b1 | 826 | 0.536 | -0.0066 | No |
| 3 | Pex6 | 985 | 0.495 | 0.0079 | No |
| 4 | Nudt12 | 1145 | 0.461 | 0.0204 | No |
| 5 | Isoc1 | 1245 | 0.440 | 0.0367 | No |
| 6 | Pex7 | 1491 | 0.406 | 0.0392 | No |
| 7 | Abca4 | 1678 | 0.384 | 0.0453 | No |
| 8 | Pex26 | 1914 | 0.354 | 0.0457 | No |
| 9 | Nedd4 | 1922 | 0.353 | 0.0647 | No |
| 10 | Rbp1 | 2156 | 0.322 | 0.0635 | No |
| 11 | Cyp39a1 | 2265 | 0.310 | 0.0718 | No |
| 12 | Nr3c2 | 2395 | 0.294 | 0.0776 | No |
| 13 | Abcd1 | 2746 | 0.259 | 0.0635 | No |
| 14 | Optn | 2886 | 0.244 | 0.0657 | No |
| 15 | Lonp2 | 2933 | 0.240 | 0.0752 | No |
| 16 | Abcd2 | 3005 | 0.233 | 0.0823 | No |
| 17 | Mlycd | 3025 | 0.231 | 0.0935 | No |
| 18 | Pex12 | 3029 | 0.231 | 0.1060 | No |
| 19 | Rxra | 3537 | 0.194 | 0.0755 | No |
| 20 | Aldh1a1 | 3595 | 0.189 | 0.0813 | No |
| 21 | Fdxr | 3730 | 0.178 | 0.0802 | No |
| 22 | Hsd3b7 | 3940 | 0.160 | 0.0720 | No |
| 23 | Abca1 | 4233 | 0.136 | 0.0558 | No |
| 24 | Abca5 | 4391 | 0.125 | 0.0499 | No |
| 25 | Fads2 | 4479 | 0.117 | 0.0493 | No |
| 26 | Gstk1 | 4519 | 0.114 | 0.0525 | No |
| 27 | Soat2 | 4568 | 0.110 | 0.0546 | No |
| 28 | Crot | 4721 | 0.097 | 0.0476 | No |
| 29 | Pex13 | 4908 | 0.084 | 0.0371 | No |
| 30 | Pxmp2 | 5389 | 0.048 | 0.0007 | No |
| 31 | Dio2 | 5473 | 0.043 | -0.0037 | No |
| 32 | Slc22a18 | 5590 | 0.035 | -0.0112 | No |
| 33 | Pex19 | 5682 | 0.029 | -0.0170 | No |
| 34 | Pnpla8 | 6102 | 0.000 | -0.0511 | No |
| 35 | Cyp7b1 | 6207 | -0.004 | -0.0593 | No |
| 36 | Pfkm | 6231 | -0.006 | -0.0608 | No |
| 37 | Hacl1 | 6292 | -0.010 | -0.0651 | No |
| 38 | Pex16 | 6295 | -0.011 | -0.0647 | No |
| 39 | Prdx5 | 6495 | -0.025 | -0.0795 | No |
| 40 | Lck | 7025 | -0.061 | -0.1192 | No |
| 41 | Aqp9 | 7077 | -0.064 | -0.1198 | No |
| 42 | Cat | 7096 | -0.065 | -0.1177 | No |
| 43 | Phyh | 7280 | -0.076 | -0.1284 | No |
| 44 | Fads1 | 7770 | -0.109 | -0.1622 | No |
| 45 | Pecr | 7925 | -0.118 | -0.1681 | No |
| 46 | Acsl5 | 8007 | -0.123 | -0.1679 | No |
| 47 | Pex11a | 8189 | -0.137 | -0.1751 | No |
| 48 | Abcd3 | 8205 | -0.138 | -0.1687 | No |
| 49 | Gclm | 8300 | -0.145 | -0.1683 | No |
| 50 | Dhcr24 | 8341 | -0.148 | -0.1634 | No |
| 51 | Abca2 | 8362 | -0.149 | -0.1568 | No |
| 52 | Gnpat | 8701 | -0.172 | -0.1748 | No |
| 53 | Aldh9a1 | 8837 | -0.183 | -0.1757 | No |
| 54 | Scp2 | 9043 | -0.198 | -0.1814 | No |
| 55 | Npc1 | 9320 | -0.219 | -0.1917 | No |
| 56 | Pex1 | 9321 | -0.219 | -0.1796 | No |
| 57 | Ar | 9840 | -0.262 | -0.2073 | No |
| 58 | Sod1 | 9993 | -0.277 | -0.2044 | No |
| 59 | Cyp27a1 | 10173 | -0.293 | -0.2027 | No |
| 60 | Slc35b2 | 10174 | -0.293 | -0.1865 | No |
| 61 | Slc29a1 | 10716 | -0.348 | -0.2113 | No |
| 62 | Paox | 10992 | -0.386 | -0.2123 | Yes |
| 63 | Pex11g | 11017 | -0.389 | -0.1927 | Yes |
| 64 | Bcar3 | 11044 | -0.393 | -0.1731 | Yes |
| 65 | Hsd17b11 | 11162 | -0.411 | -0.1599 | Yes |
| 66 | Amacr | 11190 | -0.416 | -0.1392 | Yes |
| 67 | Abca3 | 11199 | -0.416 | -0.1168 | Yes |
| 68 | Retsat | 11487 | -0.466 | -0.1144 | Yes |
| 69 | Acsl1 | 11488 | -0.466 | -0.0886 | Yes |
| 70 | Lipe | 11556 | -0.480 | -0.0676 | Yes |
| 71 | Idh2 | 11920 | -0.565 | -0.0659 | Yes |
| 72 | Idh1 | 11926 | -0.566 | -0.0350 | Yes |
| 73 | Hsd17b4 | 12028 | -0.604 | -0.0098 | Yes |
| 74 | Atxn1 | 12149 | -0.669 | 0.0174 | Yes |