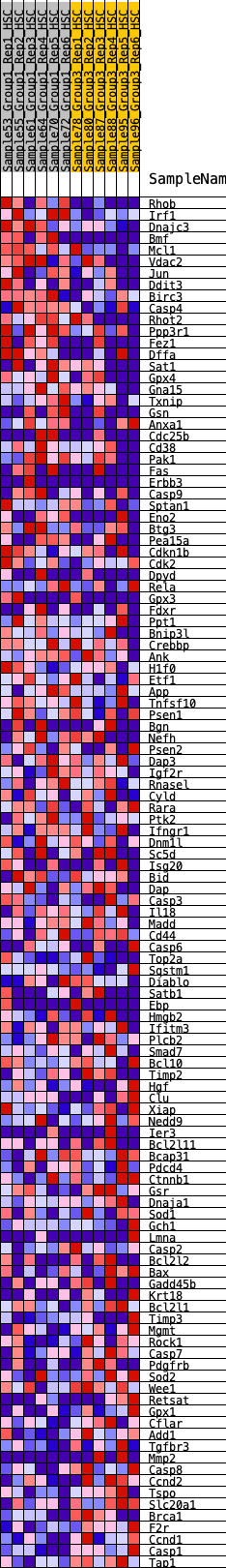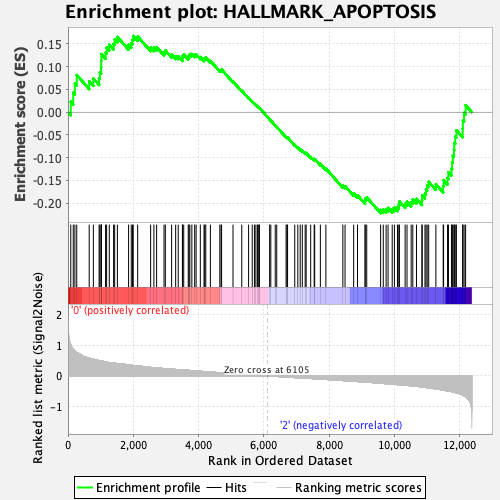
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group3.HSC_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_APOPTOSIS |
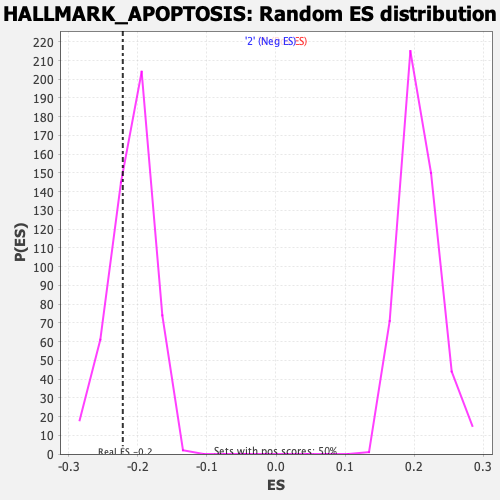
| Enrichment Score (ES) | -0.22176616 |
| Normalized Enrichment Score (NES) | -1.060242 |
| Nominal p-value | 0.31150794 |
| FDR q-value | 0.7743076 |
| FWER p-Value | 0.996 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rhob | 85 | 1.009 | 0.0231 | No |
| 2 | Irf1 | 161 | 0.874 | 0.0431 | No |
| 3 | Dnajc3 | 211 | 0.816 | 0.0634 | No |
| 4 | Bmf | 268 | 0.769 | 0.0817 | No |
| 5 | Mcl1 | 649 | 0.579 | 0.0680 | No |
| 6 | Vdac2 | 776 | 0.548 | 0.0740 | No |
| 7 | Jun | 952 | 0.502 | 0.0747 | No |
| 8 | Ddit3 | 975 | 0.497 | 0.0877 | No |
| 9 | Birc3 | 1014 | 0.490 | 0.0992 | No |
| 10 | Casp4 | 1018 | 0.489 | 0.1135 | No |
| 11 | Rhot2 | 1022 | 0.486 | 0.1278 | No |
| 12 | Ppp3r1 | 1151 | 0.460 | 0.1310 | No |
| 13 | Fez1 | 1182 | 0.454 | 0.1421 | No |
| 14 | Dffa | 1264 | 0.435 | 0.1485 | No |
| 15 | Sat1 | 1396 | 0.417 | 0.1502 | No |
| 16 | Gpx4 | 1429 | 0.413 | 0.1599 | No |
| 17 | Gna15 | 1512 | 0.403 | 0.1653 | No |
| 18 | Txnip | 1855 | 0.363 | 0.1482 | No |
| 19 | Gsn | 1937 | 0.350 | 0.1520 | No |
| 20 | Anxa1 | 1975 | 0.345 | 0.1592 | No |
| 21 | Cdc25b | 1997 | 0.342 | 0.1677 | No |
| 22 | Cd38 | 2133 | 0.324 | 0.1663 | No |
| 23 | Pak1 | 2530 | 0.277 | 0.1422 | No |
| 24 | Fas | 2630 | 0.269 | 0.1422 | No |
| 25 | Erbb3 | 2713 | 0.261 | 0.1433 | No |
| 26 | Casp9 | 2940 | 0.239 | 0.1319 | No |
| 27 | Sptan1 | 2980 | 0.234 | 0.1357 | No |
| 28 | Eno2 | 3173 | 0.221 | 0.1266 | No |
| 29 | Btg3 | 3296 | 0.210 | 0.1229 | No |
| 30 | Pea15a | 3374 | 0.205 | 0.1228 | No |
| 31 | Cdkn1b | 3503 | 0.197 | 0.1182 | No |
| 32 | Cdk2 | 3508 | 0.196 | 0.1237 | No |
| 33 | Dpyd | 3543 | 0.194 | 0.1267 | No |
| 34 | Rela | 3681 | 0.182 | 0.1209 | No |
| 35 | Gpx3 | 3704 | 0.180 | 0.1245 | No |
| 36 | Fdxr | 3730 | 0.178 | 0.1278 | No |
| 37 | Ppt1 | 3792 | 0.173 | 0.1279 | No |
| 38 | Bnip3l | 3878 | 0.165 | 0.1259 | No |
| 39 | Crebbp | 3922 | 0.161 | 0.1272 | No |
| 40 | Ank | 4051 | 0.151 | 0.1212 | No |
| 41 | H1f0 | 4165 | 0.141 | 0.1162 | No |
| 42 | Etf1 | 4186 | 0.140 | 0.1188 | No |
| 43 | App | 4218 | 0.137 | 0.1203 | No |
| 44 | Tnfsf10 | 4359 | 0.127 | 0.1127 | No |
| 45 | Psen1 | 4648 | 0.103 | 0.0922 | No |
| 46 | Bgn | 4687 | 0.099 | 0.0921 | No |
| 47 | Nefh | 4702 | 0.098 | 0.0939 | No |
| 48 | Psen2 | 5052 | 0.073 | 0.0675 | No |
| 49 | Dap3 | 5320 | 0.053 | 0.0473 | No |
| 50 | Igf2r | 5529 | 0.039 | 0.0315 | No |
| 51 | Rnasel | 5640 | 0.031 | 0.0234 | No |
| 52 | Cyld | 5708 | 0.027 | 0.0188 | No |
| 53 | Rara | 5752 | 0.024 | 0.0160 | No |
| 54 | Ptk2 | 5799 | 0.021 | 0.0128 | No |
| 55 | Ifngr1 | 5822 | 0.018 | 0.0116 | No |
| 56 | Dnm1l | 5849 | 0.017 | 0.0100 | No |
| 57 | Sc5d | 5860 | 0.015 | 0.0096 | No |
| 58 | Isg20 | 6173 | -0.002 | -0.0158 | No |
| 59 | Bid | 6209 | -0.004 | -0.0185 | No |
| 60 | Dap | 6344 | -0.014 | -0.0291 | No |
| 61 | Casp3 | 6390 | -0.017 | -0.0322 | No |
| 62 | Il18 | 6677 | -0.038 | -0.0545 | No |
| 63 | Madd | 6705 | -0.039 | -0.0555 | No |
| 64 | Cd44 | 6722 | -0.040 | -0.0556 | No |
| 65 | Casp6 | 6942 | -0.055 | -0.0718 | No |
| 66 | Top2a | 7036 | -0.061 | -0.0776 | No |
| 67 | Sqstm1 | 7109 | -0.066 | -0.0815 | No |
| 68 | Diablo | 7173 | -0.070 | -0.0846 | No |
| 69 | Satb1 | 7261 | -0.075 | -0.0895 | No |
| 70 | Ebp | 7291 | -0.077 | -0.0896 | No |
| 71 | Hmgb2 | 7430 | -0.085 | -0.0983 | No |
| 72 | Ifitm3 | 7537 | -0.092 | -0.1042 | No |
| 73 | Plcb2 | 7553 | -0.093 | -0.1026 | No |
| 74 | Smad7 | 7726 | -0.106 | -0.1135 | No |
| 75 | Bcl10 | 7895 | -0.117 | -0.1238 | No |
| 76 | Timp2 | 8415 | -0.152 | -0.1616 | No |
| 77 | Hgf | 8482 | -0.156 | -0.1623 | No |
| 78 | Clu | 8746 | -0.176 | -0.1786 | No |
| 79 | Xiap | 8863 | -0.185 | -0.1825 | No |
| 80 | Nedd9 | 9095 | -0.203 | -0.1954 | No |
| 81 | Ier3 | 9098 | -0.203 | -0.1895 | No |
| 82 | Bcl2l11 | 9146 | -0.207 | -0.1871 | No |
| 83 | Bcap31 | 9571 | -0.241 | -0.2146 | Yes |
| 84 | Pdcd4 | 9652 | -0.248 | -0.2137 | Yes |
| 85 | Ctnnb1 | 9739 | -0.255 | -0.2131 | Yes |
| 86 | Gsr | 9799 | -0.260 | -0.2102 | Yes |
| 87 | Dnaja1 | 9926 | -0.271 | -0.2124 | Yes |
| 88 | Sod1 | 9993 | -0.277 | -0.2096 | Yes |
| 89 | Gch1 | 10087 | -0.285 | -0.2086 | Yes |
| 90 | Lmna | 10128 | -0.288 | -0.2033 | Yes |
| 91 | Casp2 | 10146 | -0.290 | -0.1961 | Yes |
| 92 | Bcl2l2 | 10321 | -0.306 | -0.2012 | Yes |
| 93 | Bax | 10379 | -0.311 | -0.1966 | Yes |
| 94 | Gadd45b | 10506 | -0.326 | -0.1971 | Yes |
| 95 | Krt18 | 10556 | -0.331 | -0.1913 | Yes |
| 96 | Bcl2l1 | 10669 | -0.343 | -0.1902 | Yes |
| 97 | Timp3 | 10837 | -0.362 | -0.1930 | Yes |
| 98 | Mgmt | 10842 | -0.363 | -0.1825 | Yes |
| 99 | Rock1 | 10929 | -0.375 | -0.1784 | Yes |
| 100 | Casp7 | 10960 | -0.380 | -0.1695 | Yes |
| 101 | Pdgfrb | 10997 | -0.387 | -0.1609 | Yes |
| 102 | Sod2 | 11043 | -0.392 | -0.1529 | Yes |
| 103 | Wee1 | 11263 | -0.427 | -0.1580 | Yes |
| 104 | Retsat | 11487 | -0.466 | -0.1623 | Yes |
| 105 | Gpx1 | 11499 | -0.468 | -0.1493 | Yes |
| 106 | Cflar | 11620 | -0.492 | -0.1444 | Yes |
| 107 | Add1 | 11648 | -0.496 | -0.1318 | Yes |
| 108 | Tgfbr3 | 11740 | -0.517 | -0.1238 | Yes |
| 109 | Mmp2 | 11760 | -0.521 | -0.1099 | Yes |
| 110 | Casp8 | 11781 | -0.526 | -0.0958 | Yes |
| 111 | Ccnd2 | 11816 | -0.535 | -0.0826 | Yes |
| 112 | Tspo | 11826 | -0.538 | -0.0673 | Yes |
| 113 | Slc20a1 | 11856 | -0.547 | -0.0534 | Yes |
| 114 | Brca1 | 11891 | -0.554 | -0.0397 | Yes |
| 115 | F2r | 12083 | -0.630 | -0.0365 | Yes |
| 116 | Ccnd1 | 12089 | -0.635 | -0.0180 | Yes |
| 117 | Casp1 | 12127 | -0.661 | -0.0013 | Yes |
| 118 | Tap1 | 12173 | -0.687 | 0.0155 | Yes |