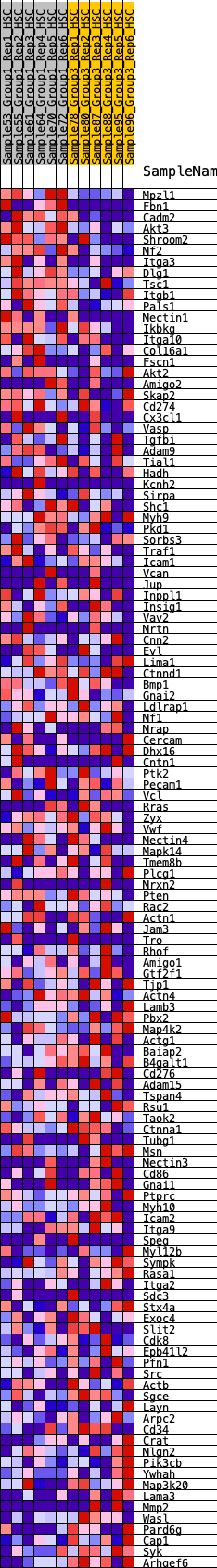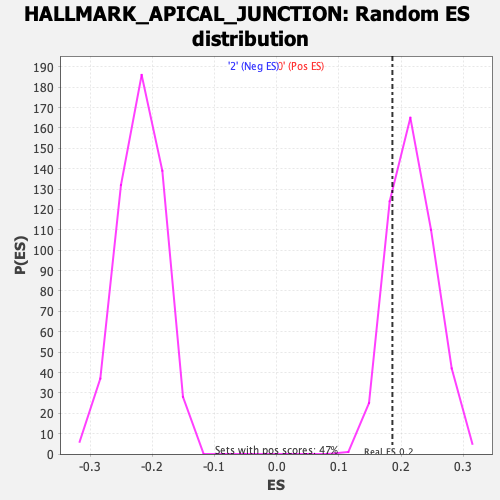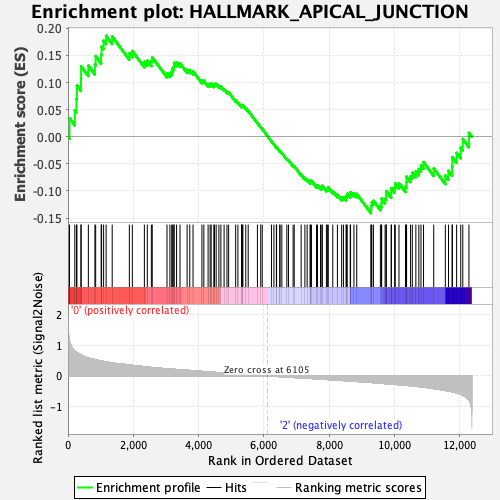
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group3.HSC_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_APICAL_JUNCTION |
| Enrichment Score (ES) | 0.18599184 |
| Normalized Enrichment Score (NES) | 0.85545087 |
| Nominal p-value | 0.7987288 |
| FDR q-value | 0.890125 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Mpzl1 | 46 | 1.143 | 0.0342 | Yes |
| 2 | Fbn1 | 208 | 0.825 | 0.0484 | Yes |
| 3 | Cadm2 | 259 | 0.774 | 0.0700 | Yes |
| 4 | Akt3 | 273 | 0.767 | 0.0944 | Yes |
| 5 | Shroom2 | 397 | 0.692 | 0.1073 | Yes |
| 6 | Nf2 | 401 | 0.691 | 0.1300 | Yes |
| 7 | Itga3 | 623 | 0.585 | 0.1314 | Yes |
| 8 | Dlg1 | 824 | 0.537 | 0.1328 | Yes |
| 9 | Tsc1 | 847 | 0.529 | 0.1486 | Yes |
| 10 | Itgb1 | 1015 | 0.490 | 0.1512 | Yes |
| 11 | Pals1 | 1028 | 0.484 | 0.1663 | Yes |
| 12 | Nectin1 | 1090 | 0.473 | 0.1770 | Yes |
| 13 | Ikbkg | 1167 | 0.457 | 0.1860 | Yes |
| 14 | Itga10 | 1353 | 0.423 | 0.1849 | No |
| 15 | Col16a1 | 1878 | 0.360 | 0.1541 | No |
| 16 | Fscn1 | 1970 | 0.345 | 0.1581 | No |
| 17 | Akt2 | 2338 | 0.301 | 0.1381 | No |
| 18 | Amigo2 | 2428 | 0.289 | 0.1404 | No |
| 19 | Skap2 | 2552 | 0.276 | 0.1395 | No |
| 20 | Cd274 | 2579 | 0.274 | 0.1465 | No |
| 21 | Cx3cl1 | 3031 | 0.231 | 0.1173 | No |
| 22 | Vasp | 3120 | 0.225 | 0.1176 | No |
| 23 | Tgfbi | 3176 | 0.221 | 0.1204 | No |
| 24 | Adam9 | 3204 | 0.219 | 0.1255 | No |
| 25 | Tial1 | 3241 | 0.215 | 0.1297 | No |
| 26 | Hadh | 3249 | 0.215 | 0.1363 | No |
| 27 | Kcnh2 | 3324 | 0.209 | 0.1371 | No |
| 28 | Sirpa | 3429 | 0.202 | 0.1354 | No |
| 29 | Shc1 | 3647 | 0.186 | 0.1238 | No |
| 30 | Myh9 | 3728 | 0.178 | 0.1232 | No |
| 31 | Pkd1 | 3828 | 0.169 | 0.1207 | No |
| 32 | Sorbs3 | 4100 | 0.146 | 0.1034 | No |
| 33 | Traf1 | 4159 | 0.142 | 0.1034 | No |
| 34 | Icam1 | 4289 | 0.132 | 0.0972 | No |
| 35 | Vcan | 4351 | 0.127 | 0.0964 | No |
| 36 | Jup | 4383 | 0.125 | 0.0981 | No |
| 37 | Inppl1 | 4465 | 0.118 | 0.0954 | No |
| 38 | Insig1 | 4485 | 0.117 | 0.0977 | No |
| 39 | Vav2 | 4536 | 0.112 | 0.0973 | No |
| 40 | Nrtn | 4620 | 0.105 | 0.0941 | No |
| 41 | Cnn2 | 4681 | 0.100 | 0.0925 | No |
| 42 | Evl | 4779 | 0.093 | 0.0876 | No |
| 43 | Lima1 | 4867 | 0.086 | 0.0834 | No |
| 44 | Ctnnd1 | 4918 | 0.083 | 0.0821 | No |
| 45 | Bmp1 | 5133 | 0.066 | 0.0668 | No |
| 46 | Gnai2 | 5202 | 0.061 | 0.0632 | No |
| 47 | Ldlrap1 | 5312 | 0.054 | 0.0561 | No |
| 48 | Nf1 | 5313 | 0.054 | 0.0579 | No |
| 49 | Nrap | 5347 | 0.052 | 0.0569 | No |
| 50 | Cercam | 5351 | 0.051 | 0.0584 | No |
| 51 | Dhx16 | 5443 | 0.045 | 0.0525 | No |
| 52 | Cntn1 | 5517 | 0.040 | 0.0478 | No |
| 53 | Ptk2 | 5799 | 0.021 | 0.0256 | No |
| 54 | Pecam1 | 5901 | 0.012 | 0.0177 | No |
| 55 | Vcl | 5947 | 0.010 | 0.0144 | No |
| 56 | Rras | 6229 | -0.006 | -0.0084 | No |
| 57 | Zyx | 6304 | -0.011 | -0.0141 | No |
| 58 | Vwf | 6379 | -0.016 | -0.0196 | No |
| 59 | Nectin4 | 6386 | -0.017 | -0.0195 | No |
| 60 | Mapk14 | 6484 | -0.025 | -0.0266 | No |
| 61 | Tmem8b | 6490 | -0.025 | -0.0262 | No |
| 62 | Plcg1 | 6547 | -0.029 | -0.0298 | No |
| 63 | Nrxn2 | 6700 | -0.039 | -0.0410 | No |
| 64 | Pten | 6750 | -0.042 | -0.0436 | No |
| 65 | Rac2 | 6892 | -0.052 | -0.0534 | No |
| 66 | Actn1 | 6926 | -0.054 | -0.0543 | No |
| 67 | Jam3 | 7138 | -0.068 | -0.0693 | No |
| 68 | Tro | 7253 | -0.075 | -0.0761 | No |
| 69 | Rhof | 7322 | -0.078 | -0.0790 | No |
| 70 | Amigo1 | 7414 | -0.084 | -0.0837 | No |
| 71 | Gtf2f1 | 7420 | -0.084 | -0.0813 | No |
| 72 | Tjp1 | 7453 | -0.086 | -0.0811 | No |
| 73 | Actn4 | 7609 | -0.099 | -0.0905 | No |
| 74 | Lamb3 | 7638 | -0.101 | -0.0894 | No |
| 75 | Pbx2 | 7731 | -0.106 | -0.0934 | No |
| 76 | Map4k2 | 7774 | -0.109 | -0.0932 | No |
| 77 | Actg1 | 7786 | -0.111 | -0.0904 | No |
| 78 | Baiap2 | 7911 | -0.118 | -0.0966 | No |
| 79 | B4galt1 | 7949 | -0.120 | -0.0957 | No |
| 80 | Cd276 | 7968 | -0.122 | -0.0931 | No |
| 81 | Adam15 | 8107 | -0.131 | -0.1000 | No |
| 82 | Tspan4 | 8248 | -0.141 | -0.1068 | No |
| 83 | Rsu1 | 8378 | -0.150 | -0.1123 | No |
| 84 | Taok2 | 8433 | -0.153 | -0.1117 | No |
| 85 | Ctnna1 | 8514 | -0.158 | -0.1130 | No |
| 86 | Tubg1 | 8525 | -0.159 | -0.1085 | No |
| 87 | Msn | 8549 | -0.161 | -0.1050 | No |
| 88 | Nectin3 | 8640 | -0.167 | -0.1068 | No |
| 89 | Cd86 | 8651 | -0.168 | -0.1021 | No |
| 90 | Gnai1 | 8754 | -0.177 | -0.1045 | No |
| 91 | Ptprc | 8840 | -0.183 | -0.1054 | No |
| 92 | Myh10 | 9275 | -0.216 | -0.1337 | No |
| 93 | Icam2 | 9283 | -0.216 | -0.1271 | No |
| 94 | Itga9 | 9301 | -0.217 | -0.1213 | No |
| 95 | Speg | 9351 | -0.222 | -0.1179 | No |
| 96 | Myl12b | 9564 | -0.240 | -0.1273 | No |
| 97 | Sympk | 9600 | -0.243 | -0.1221 | No |
| 98 | Rasa1 | 9602 | -0.243 | -0.1141 | No |
| 99 | Itga2 | 9708 | -0.253 | -0.1143 | No |
| 100 | Sdc3 | 9741 | -0.255 | -0.1084 | No |
| 101 | Stx4a | 9744 | -0.256 | -0.1001 | No |
| 102 | Exoc4 | 9897 | -0.268 | -0.1036 | No |
| 103 | Slit2 | 9898 | -0.268 | -0.0947 | No |
| 104 | Cdk8 | 10000 | -0.277 | -0.0938 | No |
| 105 | Epb41l2 | 10018 | -0.279 | -0.0859 | No |
| 106 | Pfn1 | 10133 | -0.288 | -0.0856 | No |
| 107 | Src | 10333 | -0.307 | -0.0917 | No |
| 108 | Actb | 10363 | -0.310 | -0.0838 | No |
| 109 | Sgce | 10366 | -0.310 | -0.0737 | No |
| 110 | Layn | 10488 | -0.324 | -0.0728 | No |
| 111 | Arpc2 | 10541 | -0.329 | -0.0661 | No |
| 112 | Cd34 | 10650 | -0.341 | -0.0636 | No |
| 113 | Crat | 10742 | -0.351 | -0.0594 | No |
| 114 | Nlgn2 | 10807 | -0.359 | -0.0527 | No |
| 115 | Pik3cb | 10885 | -0.368 | -0.0468 | No |
| 116 | Ywhah | 11197 | -0.416 | -0.0584 | No |
| 117 | Map3k20 | 11552 | -0.479 | -0.0714 | No |
| 118 | Lama3 | 11650 | -0.497 | -0.0629 | No |
| 119 | Mmp2 | 11760 | -0.521 | -0.0545 | No |
| 120 | Wasl | 11767 | -0.524 | -0.0376 | No |
| 121 | Pard6g | 11898 | -0.557 | -0.0297 | No |
| 122 | Cap1 | 12024 | -0.604 | -0.0199 | No |
| 123 | Syk | 12087 | -0.633 | -0.0040 | No |
| 124 | Arhgef6 | 12275 | -0.797 | 0.0072 | No |