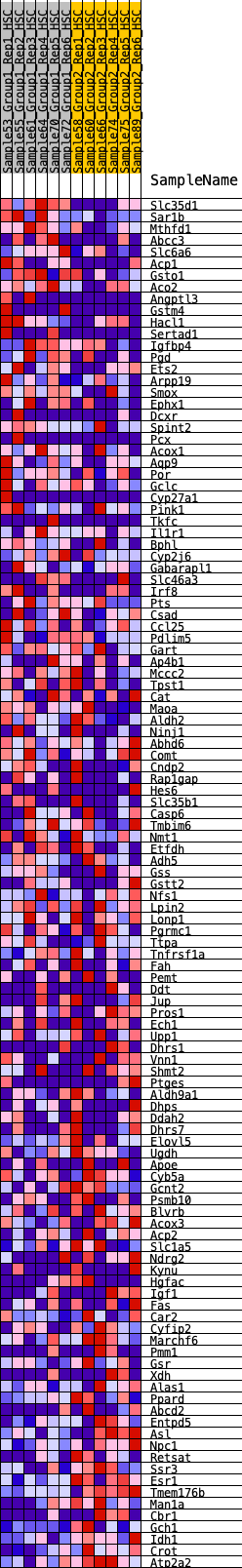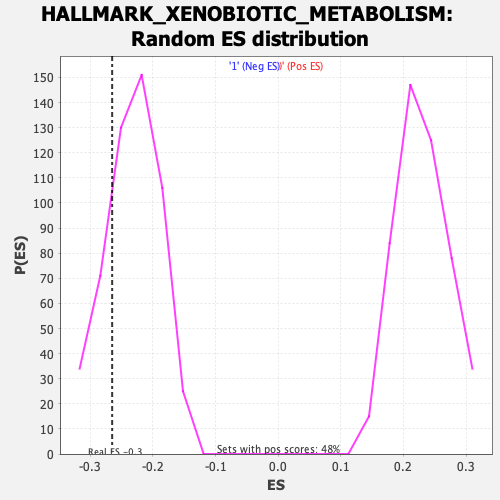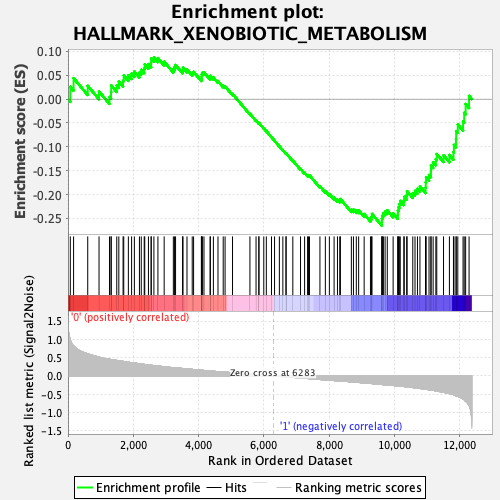
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group2.HSC_Pheno.cls#Group1_versus_Group2_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_XENOBIOTIC_METABOLISM |
| Enrichment Score (ES) | -0.2650316 |
| Normalized Enrichment Score (NES) | -1.1437984 |
| Nominal p-value | 0.21276596 |
| FDR q-value | 0.48690936 |
| FWER p-Value | 0.951 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Slc35d1 | 71 | 0.999 | 0.0259 | No |
| 2 | Sar1b | 171 | 0.823 | 0.0440 | No |
| 3 | Mthfd1 | 604 | 0.605 | 0.0279 | No |
| 4 | Abcc3 | 950 | 0.514 | 0.0161 | No |
| 5 | Slc6a6 | 1270 | 0.456 | 0.0045 | No |
| 6 | Acp1 | 1318 | 0.449 | 0.0149 | No |
| 7 | Gsto1 | 1323 | 0.449 | 0.0289 | No |
| 8 | Aco2 | 1493 | 0.424 | 0.0285 | No |
| 9 | Angptl3 | 1556 | 0.415 | 0.0366 | No |
| 10 | Gstm4 | 1686 | 0.397 | 0.0387 | No |
| 11 | Hacl1 | 1707 | 0.395 | 0.0496 | No |
| 12 | Sertad1 | 1848 | 0.375 | 0.0501 | No |
| 13 | Igfbp4 | 1947 | 0.361 | 0.0536 | No |
| 14 | Pgd | 2032 | 0.350 | 0.0578 | No |
| 15 | Ets2 | 2187 | 0.333 | 0.0558 | No |
| 16 | Arpp19 | 2243 | 0.325 | 0.0617 | No |
| 17 | Smox | 2333 | 0.316 | 0.0644 | No |
| 18 | Ephx1 | 2350 | 0.314 | 0.0731 | No |
| 19 | Dcxr | 2463 | 0.301 | 0.0735 | No |
| 20 | Spint2 | 2546 | 0.290 | 0.0760 | No |
| 21 | Pcx | 2547 | 0.290 | 0.0852 | No |
| 22 | Acox1 | 2630 | 0.282 | 0.0875 | No |
| 23 | Aqp9 | 2753 | 0.270 | 0.0861 | No |
| 24 | Por | 2944 | 0.252 | 0.0786 | No |
| 25 | Gclc | 3225 | 0.226 | 0.0629 | No |
| 26 | Cyp27a1 | 3262 | 0.224 | 0.0671 | No |
| 27 | Pink1 | 3293 | 0.222 | 0.0717 | No |
| 28 | Tkfc | 3507 | 0.204 | 0.0607 | No |
| 29 | Il1r1 | 3521 | 0.203 | 0.0661 | No |
| 30 | Bphl | 3643 | 0.192 | 0.0623 | No |
| 31 | Cyp2j6 | 3803 | 0.179 | 0.0550 | No |
| 32 | Gabarapl1 | 3845 | 0.176 | 0.0573 | No |
| 33 | Slc46a3 | 4083 | 0.156 | 0.0429 | No |
| 34 | Irf8 | 4091 | 0.156 | 0.0473 | No |
| 35 | Pts | 4094 | 0.155 | 0.0520 | No |
| 36 | Csad | 4114 | 0.153 | 0.0554 | No |
| 37 | Ccl25 | 4163 | 0.150 | 0.0562 | No |
| 38 | Pdlim5 | 4351 | 0.134 | 0.0452 | No |
| 39 | Gart | 4358 | 0.134 | 0.0489 | No |
| 40 | Ap4b1 | 4451 | 0.128 | 0.0455 | No |
| 41 | Mccc2 | 4592 | 0.118 | 0.0378 | No |
| 42 | Tpst1 | 4752 | 0.106 | 0.0282 | No |
| 43 | Cat | 4809 | 0.103 | 0.0269 | No |
| 44 | Maoa | 5037 | 0.087 | 0.0111 | No |
| 45 | Aldh2 | 5570 | 0.051 | -0.0307 | No |
| 46 | Ninj1 | 5757 | 0.038 | -0.0447 | No |
| 47 | Abhd6 | 5843 | 0.032 | -0.0506 | No |
| 48 | Comt | 5846 | 0.031 | -0.0498 | No |
| 49 | Cndp2 | 5994 | 0.020 | -0.0612 | No |
| 50 | Rap1gap | 6072 | 0.014 | -0.0670 | No |
| 51 | Hes6 | 6230 | 0.004 | -0.0797 | No |
| 52 | Slc35b1 | 6324 | -0.001 | -0.0872 | No |
| 53 | Casp6 | 6471 | -0.011 | -0.0988 | No |
| 54 | Tmbim6 | 6578 | -0.018 | -0.1069 | No |
| 55 | Nmt1 | 6675 | -0.023 | -0.1140 | No |
| 56 | Etfdh | 6676 | -0.023 | -0.1132 | No |
| 57 | Adh5 | 6883 | -0.039 | -0.1288 | No |
| 58 | Gss | 7119 | -0.055 | -0.1463 | No |
| 59 | Gstt2 | 7244 | -0.062 | -0.1544 | No |
| 60 | Nfs1 | 7335 | -0.069 | -0.1595 | No |
| 61 | Lpin2 | 7362 | -0.071 | -0.1594 | No |
| 62 | Lonp1 | 7393 | -0.073 | -0.1596 | No |
| 63 | Pgrmc1 | 7713 | -0.096 | -0.1826 | No |
| 64 | Ttpa | 7879 | -0.108 | -0.1926 | No |
| 65 | Tnfrsf1a | 8002 | -0.115 | -0.1989 | No |
| 66 | Fah | 8147 | -0.125 | -0.2067 | No |
| 67 | Pemt | 8250 | -0.132 | -0.2109 | No |
| 68 | Ddt | 8320 | -0.137 | -0.2122 | No |
| 69 | Jup | 8340 | -0.138 | -0.2093 | No |
| 70 | Pros1 | 8675 | -0.159 | -0.2315 | No |
| 71 | Ech1 | 8739 | -0.165 | -0.2314 | No |
| 72 | Upp1 | 8827 | -0.173 | -0.2330 | No |
| 73 | Dhrs1 | 8898 | -0.178 | -0.2331 | No |
| 74 | Vnn1 | 9067 | -0.190 | -0.2408 | No |
| 75 | Shmt2 | 9260 | -0.203 | -0.2500 | No |
| 76 | Ptges | 9293 | -0.206 | -0.2461 | No |
| 77 | Aldh9a1 | 9307 | -0.208 | -0.2405 | No |
| 78 | Dhps | 9608 | -0.233 | -0.2576 | Yes |
| 79 | Ddah2 | 9610 | -0.233 | -0.2503 | Yes |
| 80 | Dhrs7 | 9626 | -0.235 | -0.2441 | Yes |
| 81 | Elovl5 | 9656 | -0.237 | -0.2389 | Yes |
| 82 | Ugdh | 9708 | -0.242 | -0.2354 | Yes |
| 83 | Apoe | 9775 | -0.246 | -0.2330 | Yes |
| 84 | Cyb5a | 9954 | -0.260 | -0.2392 | Yes |
| 85 | Gcnt2 | 10097 | -0.270 | -0.2423 | Yes |
| 86 | Psmb10 | 10100 | -0.270 | -0.2338 | Yes |
| 87 | Blvrb | 10119 | -0.272 | -0.2267 | Yes |
| 88 | Acox3 | 10145 | -0.275 | -0.2200 | Yes |
| 89 | Acp2 | 10172 | -0.278 | -0.2133 | Yes |
| 90 | Slc1a5 | 10282 | -0.288 | -0.2130 | Yes |
| 91 | Ndrg2 | 10294 | -0.289 | -0.2047 | Yes |
| 92 | Kynu | 10370 | -0.296 | -0.2015 | Yes |
| 93 | Hgfac | 10381 | -0.298 | -0.1928 | Yes |
| 94 | Igf1 | 10556 | -0.316 | -0.1970 | Yes |
| 95 | Fas | 10624 | -0.323 | -0.1922 | Yes |
| 96 | Car2 | 10699 | -0.331 | -0.1877 | Yes |
| 97 | Cyfip2 | 10775 | -0.341 | -0.1830 | Yes |
| 98 | Marchf6 | 10946 | -0.361 | -0.1854 | Yes |
| 99 | Pmm1 | 10952 | -0.362 | -0.1743 | Yes |
| 100 | Gsr | 10963 | -0.363 | -0.1636 | Yes |
| 101 | Xdh | 11053 | -0.377 | -0.1589 | Yes |
| 102 | Alas1 | 11112 | -0.385 | -0.1514 | Yes |
| 103 | Ppard | 11114 | -0.385 | -0.1392 | Yes |
| 104 | Abcd2 | 11179 | -0.393 | -0.1320 | Yes |
| 105 | Entpd5 | 11262 | -0.406 | -0.1258 | Yes |
| 106 | Asl | 11290 | -0.410 | -0.1150 | Yes |
| 107 | Npc1 | 11497 | -0.446 | -0.1176 | Yes |
| 108 | Retsat | 11677 | -0.480 | -0.1170 | Yes |
| 109 | Ssr3 | 11801 | -0.508 | -0.1109 | Yes |
| 110 | Esr1 | 11816 | -0.511 | -0.0958 | Yes |
| 111 | Tmem176b | 11880 | -0.539 | -0.0839 | Yes |
| 112 | Man1a | 11885 | -0.541 | -0.0670 | Yes |
| 113 | Cbr1 | 11934 | -0.557 | -0.0532 | Yes |
| 114 | Gch1 | 12095 | -0.626 | -0.0464 | Yes |
| 115 | Idh1 | 12130 | -0.650 | -0.0286 | Yes |
| 116 | Crot | 12174 | -0.690 | -0.0102 | Yes |
| 117 | Atp2a2 | 12279 | -0.804 | 0.0069 | Yes |