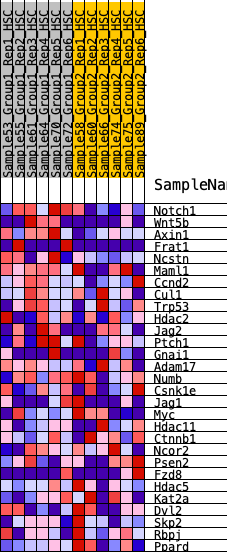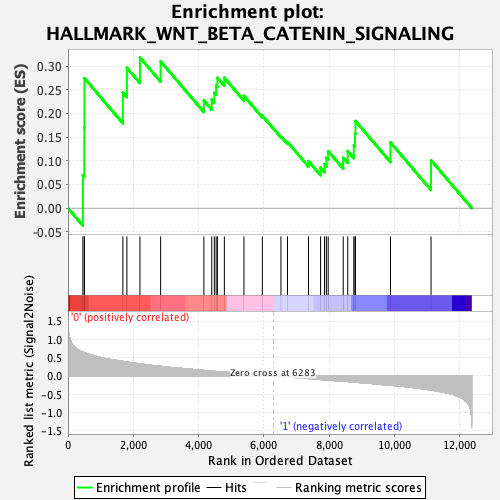
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group2.HSC_Pheno.cls#Group1_versus_Group2_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_WNT_BETA_CATENIN_SIGNALING |
| Enrichment Score (ES) | 0.3183596 |
| Normalized Enrichment Score (NES) | 1.0678366 |
| Nominal p-value | 0.31791908 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.991 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Notch1 | 455 | 0.652 | 0.0698 | Yes |
| 2 | Wnt5b | 499 | 0.638 | 0.1706 | Yes |
| 3 | Axin1 | 504 | 0.637 | 0.2745 | Yes |
| 4 | Frat1 | 1680 | 0.398 | 0.2444 | Yes |
| 5 | Ncstn | 1803 | 0.380 | 0.2966 | Yes |
| 6 | Maml1 | 2203 | 0.331 | 0.3184 | Yes |
| 7 | Ccnd2 | 2836 | 0.263 | 0.3101 | No |
| 8 | Cul1 | 4160 | 0.150 | 0.2274 | No |
| 9 | Trp53 | 4401 | 0.131 | 0.2294 | No |
| 10 | Hdac2 | 4480 | 0.126 | 0.2436 | No |
| 11 | Jag2 | 4535 | 0.122 | 0.2592 | No |
| 12 | Ptch1 | 4574 | 0.119 | 0.2756 | No |
| 13 | Gnai1 | 4787 | 0.104 | 0.2755 | No |
| 14 | Adam17 | 5387 | 0.063 | 0.2373 | No |
| 15 | Numb | 5950 | 0.023 | 0.1954 | No |
| 16 | Csnk1e | 6520 | -0.014 | 0.1516 | No |
| 17 | Jag1 | 6720 | -0.027 | 0.1398 | No |
| 18 | Myc | 7363 | -0.071 | 0.0994 | No |
| 19 | Hdac11 | 7733 | -0.097 | 0.0854 | No |
| 20 | Ctnnb1 | 7854 | -0.105 | 0.0929 | No |
| 21 | Ncor2 | 7910 | -0.110 | 0.1065 | No |
| 22 | Psen2 | 7966 | -0.112 | 0.1204 | No |
| 23 | Fzd8 | 8426 | -0.144 | 0.1067 | No |
| 24 | Hdac5 | 8564 | -0.152 | 0.1205 | No |
| 25 | Kat2a | 8750 | -0.166 | 0.1327 | No |
| 26 | Dvl2 | 8786 | -0.169 | 0.1575 | No |
| 27 | Skp2 | 8801 | -0.170 | 0.1841 | No |
| 28 | Rbpj | 9873 | -0.253 | 0.1388 | No |
| 29 | Ppard | 11114 | -0.385 | 0.1013 | No |