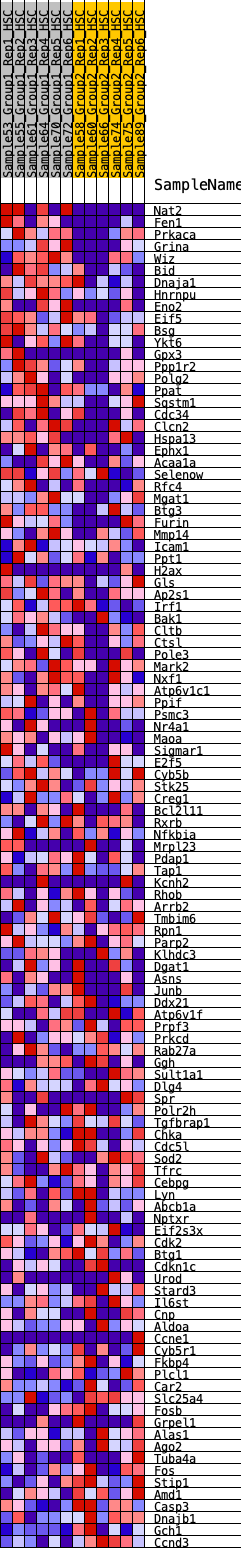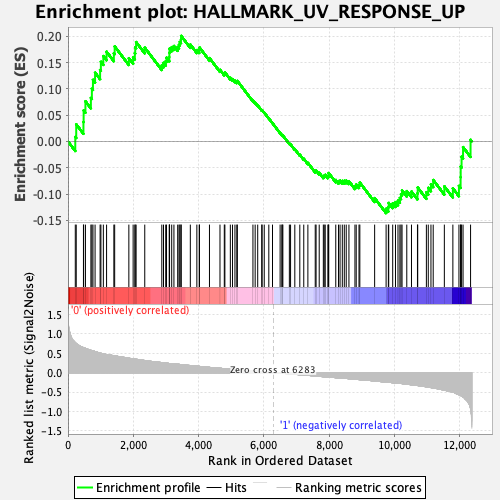
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group2.HSC_Pheno.cls#Group1_versus_Group2_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UV_RESPONSE_UP |
| Enrichment Score (ES) | 0.20055579 |
| Normalized Enrichment Score (NES) | 0.81588197 |
| Nominal p-value | 0.7653277 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nat2 | 223 | 0.774 | 0.0087 | Yes |
| 2 | Fen1 | 252 | 0.751 | 0.0325 | Yes |
| 3 | Prkaca | 473 | 0.645 | 0.0370 | Yes |
| 4 | Grina | 479 | 0.643 | 0.0589 | Yes |
| 5 | Wiz | 534 | 0.627 | 0.0763 | Yes |
| 6 | Bid | 701 | 0.576 | 0.0828 | Yes |
| 7 | Dnaja1 | 731 | 0.570 | 0.1002 | Yes |
| 8 | Hnrnpu | 764 | 0.561 | 0.1171 | Yes |
| 9 | Eno2 | 829 | 0.542 | 0.1307 | Yes |
| 10 | Eif5 | 983 | 0.506 | 0.1358 | Yes |
| 11 | Bsg | 1007 | 0.501 | 0.1514 | Yes |
| 12 | Ykt6 | 1082 | 0.486 | 0.1622 | Yes |
| 13 | Gpx3 | 1181 | 0.467 | 0.1704 | Yes |
| 14 | Ppp1r2 | 1404 | 0.435 | 0.1674 | Yes |
| 15 | Polg2 | 1429 | 0.432 | 0.1805 | Yes |
| 16 | Ppat | 1863 | 0.373 | 0.1581 | Yes |
| 17 | Sqstm1 | 1994 | 0.355 | 0.1598 | Yes |
| 18 | Cdc34 | 2047 | 0.348 | 0.1677 | Yes |
| 19 | Clcn2 | 2058 | 0.347 | 0.1790 | Yes |
| 20 | Hspa13 | 2085 | 0.345 | 0.1888 | Yes |
| 21 | Ephx1 | 2350 | 0.314 | 0.1782 | Yes |
| 22 | Acaa1a | 2872 | 0.259 | 0.1447 | Yes |
| 23 | Selenow | 2924 | 0.254 | 0.1493 | Yes |
| 24 | Rfc4 | 2992 | 0.248 | 0.1525 | Yes |
| 25 | Mgat1 | 3016 | 0.246 | 0.1591 | Yes |
| 26 | Btg3 | 3102 | 0.236 | 0.1604 | Yes |
| 27 | Furin | 3104 | 0.236 | 0.1685 | Yes |
| 28 | Mmp14 | 3109 | 0.235 | 0.1764 | Yes |
| 29 | Icam1 | 3175 | 0.230 | 0.1790 | Yes |
| 30 | Ppt1 | 3243 | 0.225 | 0.1814 | Yes |
| 31 | H2ax | 3358 | 0.217 | 0.1796 | Yes |
| 32 | Gls | 3398 | 0.214 | 0.1839 | Yes |
| 33 | Ap2s1 | 3424 | 0.212 | 0.1892 | Yes |
| 34 | Irf1 | 3459 | 0.209 | 0.1937 | Yes |
| 35 | Bak1 | 3464 | 0.208 | 0.2006 | Yes |
| 36 | Cltb | 3745 | 0.183 | 0.1841 | No |
| 37 | Ctsl | 3945 | 0.168 | 0.1737 | No |
| 38 | Pole3 | 4017 | 0.162 | 0.1735 | No |
| 39 | Mark2 | 4026 | 0.161 | 0.1784 | No |
| 40 | Nxf1 | 4331 | 0.136 | 0.1584 | No |
| 41 | Atp6v1c1 | 4653 | 0.113 | 0.1361 | No |
| 42 | Ppif | 4785 | 0.104 | 0.1290 | No |
| 43 | Psmc3 | 4803 | 0.103 | 0.1312 | No |
| 44 | Nr4a1 | 4970 | 0.093 | 0.1209 | No |
| 45 | Maoa | 5037 | 0.087 | 0.1185 | No |
| 46 | Sigmar1 | 5111 | 0.082 | 0.1154 | No |
| 47 | E2f5 | 5169 | 0.078 | 0.1135 | No |
| 48 | Cyb5b | 5184 | 0.077 | 0.1150 | No |
| 49 | Stk25 | 5662 | 0.045 | 0.0777 | No |
| 50 | Creg1 | 5730 | 0.040 | 0.0736 | No |
| 51 | Bcl2l11 | 5808 | 0.035 | 0.0685 | No |
| 52 | Rxrb | 5932 | 0.024 | 0.0593 | No |
| 53 | Nfkbia | 5942 | 0.023 | 0.0594 | No |
| 54 | Mrpl23 | 6008 | 0.019 | 0.0547 | No |
| 55 | Pdap1 | 6148 | 0.009 | 0.0437 | No |
| 56 | Tap1 | 6261 | 0.002 | 0.0346 | No |
| 57 | Kcnh2 | 6494 | -0.012 | 0.0161 | No |
| 58 | Rhob | 6547 | -0.016 | 0.0124 | No |
| 59 | Arrb2 | 6560 | -0.016 | 0.0120 | No |
| 60 | Tmbim6 | 6578 | -0.018 | 0.0112 | No |
| 61 | Rpn1 | 6777 | -0.031 | -0.0038 | No |
| 62 | Parp2 | 6814 | -0.034 | -0.0056 | No |
| 63 | Klhdc3 | 6944 | -0.043 | -0.0146 | No |
| 64 | Dgat1 | 7097 | -0.053 | -0.0252 | No |
| 65 | Asns | 7218 | -0.061 | -0.0329 | No |
| 66 | Junb | 7344 | -0.070 | -0.0407 | No |
| 67 | Ddx21 | 7569 | -0.085 | -0.0560 | No |
| 68 | Atp6v1f | 7595 | -0.087 | -0.0551 | No |
| 69 | Prpf3 | 7691 | -0.094 | -0.0595 | No |
| 70 | Prkcd | 7817 | -0.103 | -0.0662 | No |
| 71 | Rab27a | 7844 | -0.104 | -0.0646 | No |
| 72 | Ggh | 7875 | -0.107 | -0.0634 | No |
| 73 | Sult1a1 | 7956 | -0.111 | -0.0660 | No |
| 74 | Dlg4 | 7979 | -0.114 | -0.0639 | No |
| 75 | Spr | 7980 | -0.114 | -0.0599 | No |
| 76 | Polr2h | 8198 | -0.128 | -0.0732 | No |
| 77 | Tgfbrap1 | 8282 | -0.134 | -0.0753 | No |
| 78 | Chka | 8329 | -0.137 | -0.0743 | No |
| 79 | Cdc5l | 8398 | -0.142 | -0.0749 | No |
| 80 | Sod2 | 8459 | -0.146 | -0.0747 | No |
| 81 | Tfrc | 8518 | -0.150 | -0.0742 | No |
| 82 | Cebpg | 8601 | -0.155 | -0.0755 | No |
| 83 | Lyn | 8785 | -0.169 | -0.0846 | No |
| 84 | Abcb1a | 8824 | -0.173 | -0.0817 | No |
| 85 | Nptxr | 8909 | -0.178 | -0.0824 | No |
| 86 | Eif2s3x | 8935 | -0.180 | -0.0782 | No |
| 87 | Cdk2 | 9388 | -0.215 | -0.1076 | No |
| 88 | Btg1 | 9739 | -0.244 | -0.1277 | No |
| 89 | Cdkn1c | 9808 | -0.248 | -0.1246 | No |
| 90 | Urod | 9819 | -0.249 | -0.1168 | No |
| 91 | Stard3 | 9941 | -0.259 | -0.1177 | No |
| 92 | Il6st | 10026 | -0.265 | -0.1153 | No |
| 93 | Cnp | 10103 | -0.270 | -0.1121 | No |
| 94 | Aldoa | 10161 | -0.277 | -0.1072 | No |
| 95 | Ccne1 | 10195 | -0.280 | -0.1001 | No |
| 96 | Cyb5r1 | 10227 | -0.282 | -0.0929 | No |
| 97 | Fkbp4 | 10372 | -0.296 | -0.0943 | No |
| 98 | Plcl1 | 10516 | -0.312 | -0.0952 | No |
| 99 | Car2 | 10699 | -0.331 | -0.0985 | No |
| 100 | Slc25a4 | 10707 | -0.332 | -0.0875 | No |
| 101 | Fosb | 10972 | -0.364 | -0.0964 | No |
| 102 | Grpel1 | 11030 | -0.374 | -0.0881 | No |
| 103 | Alas1 | 11112 | -0.385 | -0.0813 | No |
| 104 | Ago2 | 11181 | -0.393 | -0.0732 | No |
| 105 | Tuba4a | 11522 | -0.451 | -0.0853 | No |
| 106 | Fos | 11783 | -0.502 | -0.0890 | No |
| 107 | Stip1 | 11969 | -0.570 | -0.0843 | No |
| 108 | Amd1 | 12020 | -0.588 | -0.0680 | No |
| 109 | Casp3 | 12023 | -0.589 | -0.0477 | No |
| 110 | Dnajb1 | 12050 | -0.601 | -0.0289 | No |
| 111 | Gch1 | 12095 | -0.626 | -0.0108 | No |
| 112 | Ccnd3 | 12325 | -0.936 | 0.0031 | No |