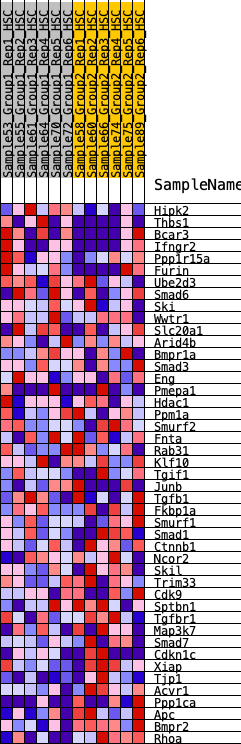
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group2.HSC_Pheno.cls#Group1_versus_Group2_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_TGF_BETA_SIGNALING |
| Enrichment Score (ES) | -0.32244942 |
| Normalized Enrichment Score (NES) | -1.2670572 |
| Nominal p-value | 0.13836478 |
| FDR q-value | 0.3736532 |
| FWER p-Value | 0.796 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hipk2 | 522 | 0.630 | 0.0216 | No |
| 2 | Thbs1 | 775 | 0.558 | 0.0579 | No |
| 3 | Bcar3 | 2232 | 0.327 | -0.0271 | No |
| 4 | Ifngr2 | 2558 | 0.289 | -0.0241 | No |
| 5 | Ppp1r15a | 2886 | 0.257 | -0.0245 | No |
| 6 | Furin | 3104 | 0.236 | -0.0182 | No |
| 7 | Ube2d3 | 3281 | 0.223 | -0.0098 | No |
| 8 | Smad6 | 3476 | 0.207 | -0.0046 | No |
| 9 | Ski | 4328 | 0.136 | -0.0598 | No |
| 10 | Wwtr1 | 4579 | 0.119 | -0.0680 | No |
| 11 | Slc20a1 | 4768 | 0.105 | -0.0726 | No |
| 12 | Arid4b | 4840 | 0.100 | -0.0681 | No |
| 13 | Bmpr1a | 5040 | 0.087 | -0.0755 | No |
| 14 | Smad3 | 5491 | 0.056 | -0.1063 | No |
| 15 | Eng | 5520 | 0.054 | -0.1031 | No |
| 16 | Pmepa1 | 5875 | 0.029 | -0.1289 | No |
| 17 | Hdac1 | 6379 | -0.006 | -0.1692 | No |
| 18 | Ppm1a | 6384 | -0.006 | -0.1689 | No |
| 19 | Smurf2 | 6473 | -0.011 | -0.1749 | No |
| 20 | Fnta | 6994 | -0.046 | -0.2125 | No |
| 21 | Rab31 | 7091 | -0.053 | -0.2149 | No |
| 22 | Klf10 | 7281 | -0.065 | -0.2237 | No |
| 23 | Tgif1 | 7283 | -0.065 | -0.2171 | No |
| 24 | Junb | 7344 | -0.070 | -0.2149 | No |
| 25 | Tgfb1 | 7357 | -0.071 | -0.2087 | No |
| 26 | Fkbp1a | 7677 | -0.093 | -0.2251 | No |
| 27 | Smurf1 | 7724 | -0.097 | -0.2190 | No |
| 28 | Smad1 | 7726 | -0.097 | -0.2093 | No |
| 29 | Ctnnb1 | 7854 | -0.105 | -0.2089 | No |
| 30 | Ncor2 | 7910 | -0.110 | -0.2022 | No |
| 31 | Skil | 8054 | -0.118 | -0.2018 | No |
| 32 | Trim33 | 8146 | -0.125 | -0.1965 | No |
| 33 | Cdk9 | 8327 | -0.137 | -0.1972 | No |
| 34 | Sptbn1 | 8694 | -0.161 | -0.2105 | No |
| 35 | Tgfbr1 | 9117 | -0.194 | -0.2251 | No |
| 36 | Map3k7 | 9151 | -0.196 | -0.2079 | No |
| 37 | Smad7 | 9734 | -0.243 | -0.2304 | No |
| 38 | Cdkn1c | 9808 | -0.248 | -0.2111 | No |
| 39 | Xiap | 10872 | -0.354 | -0.2615 | No |
| 40 | Tjp1 | 11624 | -0.470 | -0.2747 | Yes |
| 41 | Acvr1 | 11899 | -0.544 | -0.2416 | Yes |
| 42 | Ppp1ca | 12007 | -0.581 | -0.1913 | Yes |
| 43 | Apc | 12036 | -0.596 | -0.1331 | Yes |
| 44 | Bmpr2 | 12236 | -0.746 | -0.0735 | Yes |
| 45 | Rhoa | 12287 | -0.824 | 0.0062 | Yes |