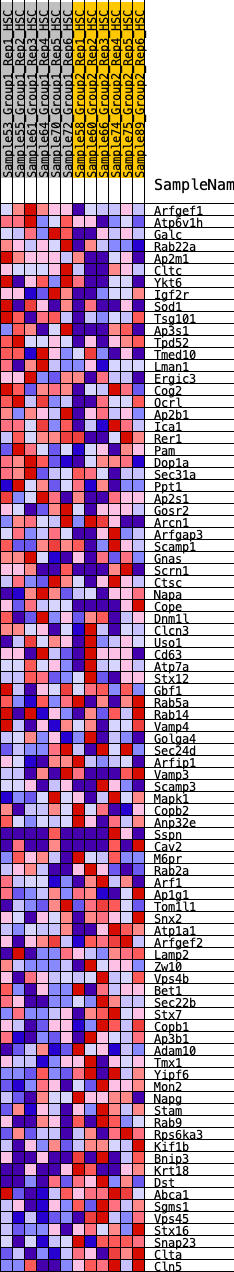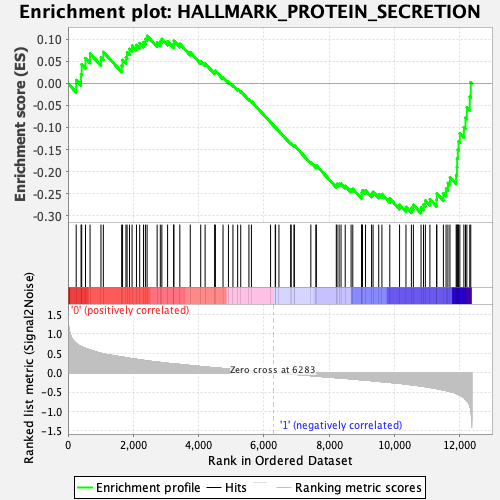
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group2.HSC_Pheno.cls#Group1_versus_Group2_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | -0.29431945 |
| Normalized Enrichment Score (NES) | -1.2890472 |
| Nominal p-value | 0.09016393 |
| FDR q-value | 0.47780886 |
| FWER p-Value | 0.763 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Arfgef1 | 251 | 0.752 | 0.0074 | No |
| 2 | Atp6v1h | 400 | 0.672 | 0.0203 | No |
| 3 | Galc | 420 | 0.662 | 0.0432 | No |
| 4 | Rab22a | 536 | 0.627 | 0.0571 | No |
| 5 | Ap2m1 | 675 | 0.585 | 0.0676 | No |
| 6 | Cltc | 1009 | 0.500 | 0.0589 | No |
| 7 | Ykt6 | 1082 | 0.486 | 0.0711 | No |
| 8 | Igf2r | 1643 | 0.404 | 0.0404 | No |
| 9 | Sod1 | 1670 | 0.399 | 0.0531 | No |
| 10 | Tsg101 | 1774 | 0.384 | 0.0590 | No |
| 11 | Ap3s1 | 1809 | 0.379 | 0.0702 | No |
| 12 | Tpd52 | 1883 | 0.370 | 0.0780 | No |
| 13 | Tmed10 | 1961 | 0.360 | 0.0851 | No |
| 14 | Lman1 | 2096 | 0.344 | 0.0869 | No |
| 15 | Ergic3 | 2195 | 0.331 | 0.0912 | No |
| 16 | Cog2 | 2312 | 0.318 | 0.0935 | No |
| 17 | Ocrl | 2370 | 0.311 | 0.1004 | No |
| 18 | Ap2b1 | 2422 | 0.306 | 0.1076 | No |
| 19 | Ica1 | 2729 | 0.273 | 0.0928 | No |
| 20 | Rer1 | 2826 | 0.264 | 0.0947 | No |
| 21 | Pam | 2873 | 0.259 | 0.1006 | No |
| 22 | Dop1a | 3049 | 0.242 | 0.0953 | No |
| 23 | Sec31a | 3233 | 0.226 | 0.0887 | No |
| 24 | Ppt1 | 3243 | 0.225 | 0.0963 | No |
| 25 | Ap2s1 | 3424 | 0.212 | 0.0895 | No |
| 26 | Gosr2 | 3743 | 0.183 | 0.0704 | No |
| 27 | Arcn1 | 4063 | 0.158 | 0.0503 | No |
| 28 | Arfgap3 | 4198 | 0.147 | 0.0448 | No |
| 29 | Scamp1 | 4488 | 0.125 | 0.0259 | No |
| 30 | Gnas | 4513 | 0.124 | 0.0285 | No |
| 31 | Scrn1 | 4746 | 0.106 | 0.0136 | No |
| 32 | Ctsc | 4912 | 0.097 | 0.0037 | No |
| 33 | Napa | 5050 | 0.086 | -0.0043 | No |
| 34 | Cope | 5194 | 0.076 | -0.0131 | No |
| 35 | Dnm1l | 5288 | 0.070 | -0.0181 | No |
| 36 | Clcn3 | 5539 | 0.053 | -0.0365 | No |
| 37 | Uso1 | 5616 | 0.048 | -0.0409 | No |
| 38 | Cd63 | 6202 | 0.006 | -0.0883 | No |
| 39 | Atp7a | 6345 | -0.003 | -0.0998 | No |
| 40 | Stx12 | 6355 | -0.004 | -0.1004 | No |
| 41 | Gbf1 | 6458 | -0.010 | -0.1083 | No |
| 42 | Rab5a | 6818 | -0.034 | -0.1363 | No |
| 43 | Rab14 | 6831 | -0.035 | -0.1360 | No |
| 44 | Vamp4 | 6920 | -0.041 | -0.1416 | No |
| 45 | Golga4 | 6929 | -0.042 | -0.1407 | No |
| 46 | Sec24d | 7435 | -0.076 | -0.1790 | No |
| 47 | Arfip1 | 7589 | -0.086 | -0.1883 | No |
| 48 | Vamp3 | 7603 | -0.088 | -0.1861 | No |
| 49 | Scamp3 | 8214 | -0.129 | -0.2310 | No |
| 50 | Mapk1 | 8236 | -0.131 | -0.2278 | No |
| 51 | Copb2 | 8300 | -0.135 | -0.2280 | No |
| 52 | Anp32e | 8359 | -0.140 | -0.2275 | No |
| 53 | Sspn | 8487 | -0.148 | -0.2324 | No |
| 54 | Cav2 | 8667 | -0.159 | -0.2411 | No |
| 55 | M6pr | 8719 | -0.163 | -0.2392 | No |
| 56 | Rab2a | 8990 | -0.185 | -0.2543 | No |
| 57 | Arf1 | 8991 | -0.185 | -0.2475 | No |
| 58 | Ap1g1 | 9016 | -0.187 | -0.2425 | No |
| 59 | Tom1l1 | 9109 | -0.193 | -0.2429 | No |
| 60 | Snx2 | 9294 | -0.206 | -0.2502 | No |
| 61 | Atp1a1 | 9340 | -0.210 | -0.2461 | No |
| 62 | Arfgef2 | 9513 | -0.225 | -0.2517 | No |
| 63 | Lamp2 | 9611 | -0.233 | -0.2510 | No |
| 64 | Zw10 | 9852 | -0.251 | -0.2612 | No |
| 65 | Vps4b | 10151 | -0.276 | -0.2753 | No |
| 66 | Bet1 | 10347 | -0.294 | -0.2803 | No |
| 67 | Sec22b | 10513 | -0.312 | -0.2822 | Yes |
| 68 | Stx7 | 10574 | -0.318 | -0.2753 | Yes |
| 69 | Copb1 | 10809 | -0.345 | -0.2815 | Yes |
| 70 | Ap3b1 | 10884 | -0.355 | -0.2744 | Yes |
| 71 | Adam10 | 10941 | -0.361 | -0.2656 | Yes |
| 72 | Tmx1 | 11080 | -0.381 | -0.2627 | Yes |
| 73 | Yipf6 | 11286 | -0.409 | -0.2642 | Yes |
| 74 | Mon2 | 11291 | -0.410 | -0.2494 | Yes |
| 75 | Napg | 11493 | -0.445 | -0.2492 | Yes |
| 76 | Stam | 11575 | -0.462 | -0.2387 | Yes |
| 77 | Rab9 | 11632 | -0.471 | -0.2258 | Yes |
| 78 | Rps6ka3 | 11694 | -0.484 | -0.2129 | Yes |
| 79 | Kif1b | 11884 | -0.541 | -0.2082 | Yes |
| 80 | Bnip3 | 11908 | -0.549 | -0.1897 | Yes |
| 81 | Krt18 | 11916 | -0.551 | -0.1699 | Yes |
| 82 | Dst | 11935 | -0.557 | -0.1507 | Yes |
| 83 | Abca1 | 11958 | -0.565 | -0.1316 | Yes |
| 84 | Sgms1 | 11997 | -0.578 | -0.1132 | Yes |
| 85 | Vps45 | 12120 | -0.641 | -0.0994 | Yes |
| 86 | Stx16 | 12170 | -0.687 | -0.0780 | Yes |
| 87 | Snap23 | 12211 | -0.720 | -0.0545 | Yes |
| 88 | Clta | 12300 | -0.851 | -0.0302 | Yes |
| 89 | Cln5 | 12331 | -0.950 | 0.0026 | Yes |