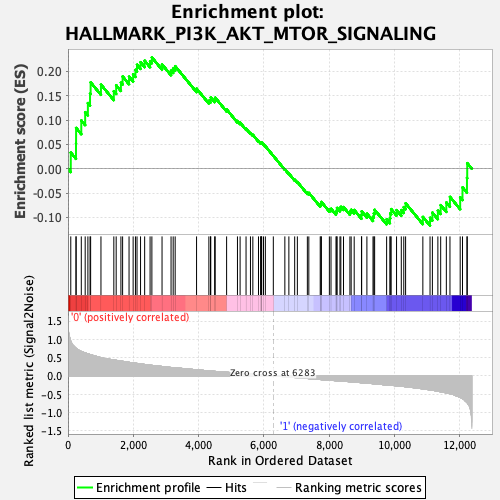
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group2.HSC_Pheno.cls#Group1_versus_Group2_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.2288587 |
| Normalized Enrichment Score (NES) | 0.9961639 |
| Nominal p-value | 0.45093945 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Raf1 | 85 | 0.950 | 0.0332 | Yes |
| 2 | Plcb1 | 245 | 0.756 | 0.0521 | Yes |
| 3 | Ube2n | 248 | 0.755 | 0.0839 | Yes |
| 4 | Traf2 | 407 | 0.669 | 0.0992 | Yes |
| 5 | Rit1 | 526 | 0.629 | 0.1162 | Yes |
| 6 | Cab39l | 609 | 0.604 | 0.1350 | Yes |
| 7 | Ap2m1 | 675 | 0.585 | 0.1544 | Yes |
| 8 | Pfn1 | 693 | 0.581 | 0.1776 | Yes |
| 9 | Cltc | 1009 | 0.500 | 0.1730 | Yes |
| 10 | Grb2 | 1403 | 0.435 | 0.1594 | Yes |
| 11 | Csnk2b | 1475 | 0.426 | 0.1716 | Yes |
| 12 | Akt1s1 | 1620 | 0.406 | 0.1770 | Yes |
| 13 | Slc2a1 | 1672 | 0.399 | 0.1897 | Yes |
| 14 | Tbk1 | 1869 | 0.372 | 0.1894 | Yes |
| 15 | Sqstm1 | 1994 | 0.355 | 0.1943 | Yes |
| 16 | Cab39 | 2065 | 0.347 | 0.2032 | Yes |
| 17 | Akt1 | 2113 | 0.342 | 0.2139 | Yes |
| 18 | Prkag1 | 2221 | 0.328 | 0.2190 | Yes |
| 19 | Il2rg | 2347 | 0.314 | 0.2221 | Yes |
| 20 | Lck | 2513 | 0.294 | 0.2211 | Yes |
| 21 | Ddit3 | 2568 | 0.289 | 0.2289 | Yes |
| 22 | Arhgdia | 2879 | 0.258 | 0.2145 | No |
| 23 | Pin1 | 3155 | 0.232 | 0.2019 | No |
| 24 | Prkar2a | 3220 | 0.227 | 0.2062 | No |
| 25 | Ube2d3 | 3281 | 0.223 | 0.2108 | No |
| 26 | Stat2 | 3936 | 0.169 | 0.1646 | No |
| 27 | Sla | 4313 | 0.138 | 0.1398 | No |
| 28 | Gsk3b | 4363 | 0.133 | 0.1414 | No |
| 29 | Il4 | 4371 | 0.133 | 0.1465 | No |
| 30 | Rac1 | 4490 | 0.125 | 0.1421 | No |
| 31 | Prkcb | 4506 | 0.124 | 0.1461 | No |
| 32 | Cdk4 | 4856 | 0.100 | 0.1219 | No |
| 33 | Smad2 | 5188 | 0.077 | 0.0982 | No |
| 34 | Myd88 | 5269 | 0.072 | 0.0947 | No |
| 35 | Ralb | 5451 | 0.059 | 0.0824 | No |
| 36 | Cdkn1b | 5586 | 0.050 | 0.0736 | No |
| 37 | Pikfyve | 5659 | 0.045 | 0.0697 | No |
| 38 | Irak4 | 5832 | 0.032 | 0.0570 | No |
| 39 | Acaca | 5899 | 0.027 | 0.0528 | No |
| 40 | Actr3 | 5912 | 0.026 | 0.0529 | No |
| 41 | Ecsit | 5916 | 0.025 | 0.0537 | No |
| 42 | Pdk1 | 5930 | 0.024 | 0.0537 | No |
| 43 | Ywhab | 5985 | 0.021 | 0.0501 | No |
| 44 | Cdk1 | 6042 | 0.017 | 0.0463 | No |
| 45 | Gna14 | 6286 | 0.000 | 0.0265 | No |
| 46 | Pten | 6639 | -0.022 | -0.0013 | No |
| 47 | Tsc2 | 6762 | -0.030 | -0.0100 | No |
| 48 | Mknk2 | 6940 | -0.043 | -0.0226 | No |
| 49 | Actr2 | 7022 | -0.047 | -0.0272 | No |
| 50 | Dusp3 | 7328 | -0.069 | -0.0491 | No |
| 51 | Vav3 | 7373 | -0.072 | -0.0497 | No |
| 52 | Dapp1 | 7722 | -0.096 | -0.0740 | No |
| 53 | Calr | 7745 | -0.098 | -0.0716 | No |
| 54 | Eif4e | 7757 | -0.099 | -0.0683 | No |
| 55 | Tnfrsf1a | 8002 | -0.115 | -0.0833 | No |
| 56 | Hsp90b1 | 8052 | -0.118 | -0.0823 | No |
| 57 | Pla2g12a | 8205 | -0.129 | -0.0893 | No |
| 58 | Pak4 | 8229 | -0.130 | -0.0857 | No |
| 59 | Mapk1 | 8236 | -0.131 | -0.0806 | No |
| 60 | Plcg1 | 8326 | -0.137 | -0.0821 | No |
| 61 | Atf1 | 8353 | -0.139 | -0.0783 | No |
| 62 | Itpr2 | 8435 | -0.144 | -0.0788 | No |
| 63 | Map2k3 | 8631 | -0.157 | -0.0881 | No |
| 64 | Ppp2r1b | 8669 | -0.159 | -0.0844 | No |
| 65 | Mapkap1 | 8762 | -0.167 | -0.0848 | No |
| 66 | Prkaa2 | 8985 | -0.184 | -0.0951 | No |
| 67 | Arf1 | 8991 | -0.185 | -0.0877 | No |
| 68 | Map3k7 | 9151 | -0.196 | -0.0924 | No |
| 69 | Mknk1 | 9338 | -0.210 | -0.0987 | No |
| 70 | Rptor | 9364 | -0.213 | -0.0918 | No |
| 71 | Cdk2 | 9388 | -0.215 | -0.0846 | No |
| 72 | Nfkbib | 9755 | -0.245 | -0.1040 | No |
| 73 | Mapk8 | 9856 | -0.252 | -0.1016 | No |
| 74 | Cxcr4 | 9867 | -0.253 | -0.0917 | No |
| 75 | Map2k6 | 9894 | -0.255 | -0.0831 | No |
| 76 | Ptpn11 | 10059 | -0.267 | -0.0851 | No |
| 77 | Arpc3 | 10204 | -0.280 | -0.0851 | No |
| 78 | E2f1 | 10280 | -0.288 | -0.0790 | No |
| 79 | Them4 | 10336 | -0.294 | -0.0711 | No |
| 80 | Trib3 | 10864 | -0.353 | -0.0991 | No |
| 81 | Cfl1 | 11084 | -0.381 | -0.1009 | No |
| 82 | Mapk9 | 11156 | -0.390 | -0.0902 | No |
| 83 | Tiam1 | 11324 | -0.416 | -0.0862 | No |
| 84 | Ripk1 | 11410 | -0.432 | -0.0749 | No |
| 85 | Nod1 | 11583 | -0.463 | -0.0694 | No |
| 86 | Rps6ka3 | 11694 | -0.484 | -0.0579 | No |
| 87 | Ppp1ca | 12007 | -0.581 | -0.0588 | No |
| 88 | Nck1 | 12078 | -0.618 | -0.0384 | No |
| 89 | Grk2 | 12215 | -0.723 | -0.0190 | No |
| 90 | Rps6ka1 | 12223 | -0.733 | 0.0114 | No |

