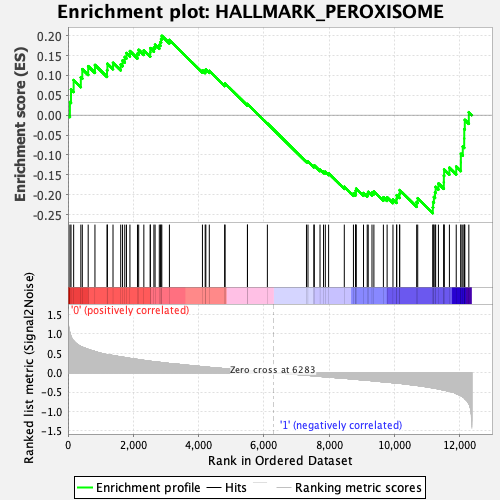
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group2.HSC_Pheno.cls#Group1_versus_Group2_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.24647467 |
| Normalized Enrichment Score (NES) | -1.1024919 |
| Nominal p-value | 0.29540917 |
| FDR q-value | 0.55655575 |
| FWER p-Value | 0.971 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sult2b1 | 59 | 1.032 | 0.0330 | No |
| 2 | Acsl5 | 92 | 0.937 | 0.0647 | No |
| 3 | Mvp | 173 | 0.821 | 0.0882 | No |
| 4 | Gstk1 | 393 | 0.674 | 0.0951 | No |
| 5 | Isoc1 | 438 | 0.658 | 0.1156 | No |
| 6 | Pex2 | 618 | 0.600 | 0.1230 | No |
| 7 | Dhrs3 | 825 | 0.545 | 0.1262 | No |
| 8 | Cacna1b | 1195 | 0.466 | 0.1132 | No |
| 9 | Ercc1 | 1207 | 0.465 | 0.1293 | No |
| 10 | Mlycd | 1378 | 0.441 | 0.1316 | No |
| 11 | Pex11b | 1614 | 0.407 | 0.1274 | No |
| 12 | Sod1 | 1670 | 0.399 | 0.1375 | No |
| 13 | Itgb1bp1 | 1738 | 0.390 | 0.1464 | No |
| 14 | Bcl10 | 1790 | 0.382 | 0.1562 | No |
| 15 | Ercc3 | 1896 | 0.368 | 0.1611 | No |
| 16 | Pex11a | 2127 | 0.341 | 0.1549 | No |
| 17 | Abcb9 | 2164 | 0.335 | 0.1642 | No |
| 18 | Ctbp1 | 2322 | 0.316 | 0.1630 | No |
| 19 | Abcc5 | 2518 | 0.294 | 0.1579 | No |
| 20 | Cln6 | 2521 | 0.293 | 0.1685 | No |
| 21 | Acox1 | 2630 | 0.282 | 0.1700 | No |
| 22 | Idh2 | 2666 | 0.279 | 0.1773 | No |
| 23 | Fdps | 2795 | 0.267 | 0.1767 | No |
| 24 | Cnbp | 2829 | 0.264 | 0.1837 | No |
| 25 | Top2a | 2850 | 0.261 | 0.1916 | No |
| 26 | Acaa1a | 2872 | 0.259 | 0.1994 | No |
| 27 | Smarcc1 | 3105 | 0.236 | 0.1891 | No |
| 28 | Ehhadh | 4116 | 0.153 | 0.1125 | No |
| 29 | Tspo | 4200 | 0.147 | 0.1111 | No |
| 30 | Ide | 4220 | 0.145 | 0.1149 | No |
| 31 | Abcd3 | 4326 | 0.137 | 0.1113 | No |
| 32 | Pex14 | 4796 | 0.104 | 0.0770 | No |
| 33 | Cat | 4809 | 0.103 | 0.0797 | No |
| 34 | Lonp2 | 5493 | 0.056 | 0.0262 | No |
| 35 | Acsl1 | 5495 | 0.056 | 0.0281 | No |
| 36 | Pex5 | 6104 | 0.012 | -0.0209 | No |
| 37 | Abcd1 | 7301 | -0.067 | -0.1158 | No |
| 38 | Nudt19 | 7347 | -0.070 | -0.1169 | No |
| 39 | Slc25a17 | 7527 | -0.081 | -0.1285 | No |
| 40 | Eci2 | 7540 | -0.082 | -0.1265 | No |
| 41 | Slc25a19 | 7716 | -0.096 | -0.1372 | No |
| 42 | Cln8 | 7823 | -0.103 | -0.1421 | No |
| 43 | Pabpc1 | 7878 | -0.108 | -0.1425 | No |
| 44 | Dlg4 | 7979 | -0.114 | -0.1465 | No |
| 45 | Sod2 | 8459 | -0.146 | -0.1802 | No |
| 46 | Ech1 | 8739 | -0.165 | -0.1969 | No |
| 47 | Acot8 | 8804 | -0.171 | -0.1958 | No |
| 48 | Hsd17b11 | 8809 | -0.171 | -0.1899 | No |
| 49 | Abcb1a | 8824 | -0.173 | -0.1847 | No |
| 50 | Dhcr24 | 9047 | -0.189 | -0.1959 | No |
| 51 | Cdk7 | 9161 | -0.197 | -0.1979 | No |
| 52 | Hmgcl | 9191 | -0.199 | -0.1930 | No |
| 53 | Aldh9a1 | 9307 | -0.208 | -0.1947 | No |
| 54 | Pex13 | 9365 | -0.213 | -0.1916 | No |
| 55 | Elovl5 | 9656 | -0.237 | -0.2065 | No |
| 56 | Prdx5 | 9770 | -0.246 | -0.2067 | No |
| 57 | Fads1 | 9949 | -0.259 | -0.2117 | No |
| 58 | Atxn1 | 10061 | -0.267 | -0.2110 | No |
| 59 | Abcb4 | 10064 | -0.268 | -0.2013 | No |
| 60 | Vps4b | 10151 | -0.276 | -0.1982 | No |
| 61 | Ywhah | 10156 | -0.276 | -0.1884 | No |
| 62 | Acsl4 | 10675 | -0.329 | -0.2186 | No |
| 63 | Slc25a4 | 10707 | -0.332 | -0.2089 | No |
| 64 | Pex6 | 11169 | -0.393 | -0.2321 | Yes |
| 65 | Abcd2 | 11179 | -0.393 | -0.2184 | Yes |
| 66 | Cadm1 | 11203 | -0.396 | -0.2058 | Yes |
| 67 | Slc23a2 | 11237 | -0.402 | -0.1938 | Yes |
| 68 | Hsd3b7 | 11256 | -0.404 | -0.1805 | Yes |
| 69 | Slc35b2 | 11341 | -0.420 | -0.1719 | Yes |
| 70 | Scp2 | 11506 | -0.448 | -0.1689 | Yes |
| 71 | Rdh11 | 11508 | -0.448 | -0.1526 | Yes |
| 72 | Aldh1a1 | 11516 | -0.450 | -0.1367 | Yes |
| 73 | Retsat | 11677 | -0.480 | -0.1321 | Yes |
| 74 | Gnpat | 11886 | -0.541 | -0.1292 | Yes |
| 75 | Fis1 | 12028 | -0.593 | -0.1190 | Yes |
| 76 | Prdx1 | 12029 | -0.594 | -0.0973 | Yes |
| 77 | Msh2 | 12087 | -0.622 | -0.0791 | Yes |
| 78 | Idh1 | 12130 | -0.650 | -0.0588 | Yes |
| 79 | Siah1a | 12132 | -0.651 | -0.0350 | Yes |
| 80 | Crat | 12149 | -0.667 | -0.0119 | Yes |
| 81 | Hsd17b4 | 12272 | -0.798 | 0.0074 | Yes |

