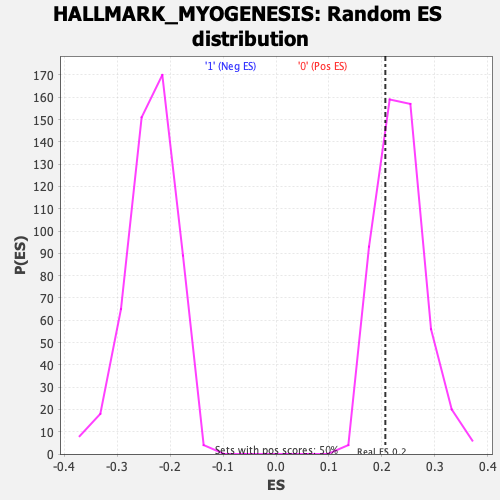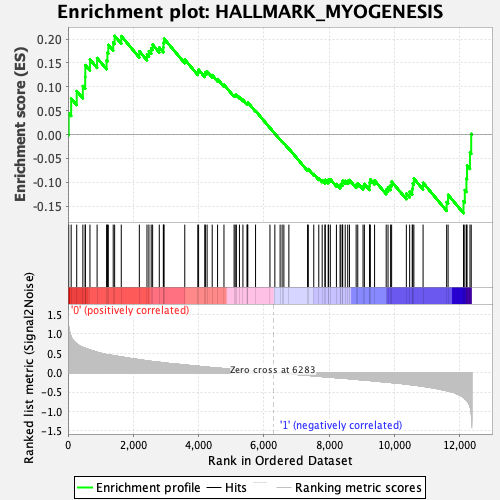
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group2.HSC_Pheno.cls#Group1_versus_Group2_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_MYOGENESIS |
| Enrichment Score (ES) | 0.20667572 |
| Normalized Enrichment Score (NES) | 0.87910235 |
| Nominal p-value | 0.6989899 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Eno3 | 22 | 1.180 | 0.0449 | Yes |
| 2 | Ocel1 | 97 | 0.925 | 0.0755 | Yes |
| 3 | Gsn | 267 | 0.742 | 0.0911 | Yes |
| 4 | Notch1 | 455 | 0.652 | 0.1017 | Yes |
| 5 | Rit1 | 526 | 0.629 | 0.1209 | Yes |
| 6 | Myom1 | 530 | 0.629 | 0.1455 | Yes |
| 7 | Mef2d | 672 | 0.586 | 0.1572 | Yes |
| 8 | Ptp4a3 | 891 | 0.527 | 0.1603 | Yes |
| 9 | Gpx3 | 1181 | 0.467 | 0.1553 | Yes |
| 10 | Aebp1 | 1208 | 0.464 | 0.1715 | Yes |
| 11 | App | 1234 | 0.461 | 0.1878 | Yes |
| 12 | Dmpk | 1384 | 0.440 | 0.1931 | Yes |
| 13 | Sorbs3 | 1428 | 0.433 | 0.2067 | Yes |
| 14 | Myh9 | 1631 | 0.405 | 0.2063 | No |
| 15 | Cnn3 | 2184 | 0.333 | 0.1745 | No |
| 16 | Foxo4 | 2418 | 0.307 | 0.1676 | No |
| 17 | Itgb1 | 2477 | 0.299 | 0.1747 | No |
| 18 | Pcx | 2547 | 0.290 | 0.1806 | No |
| 19 | Bin1 | 2591 | 0.286 | 0.1884 | No |
| 20 | Fdps | 2795 | 0.267 | 0.1824 | No |
| 21 | Mylk | 2919 | 0.255 | 0.1825 | No |
| 22 | Sh2b1 | 2923 | 0.254 | 0.1923 | No |
| 23 | Pfkm | 2942 | 0.252 | 0.2008 | No |
| 24 | Gaa | 3575 | 0.198 | 0.1572 | No |
| 25 | Akt2 | 3974 | 0.166 | 0.1313 | No |
| 26 | Pdlim7 | 3998 | 0.164 | 0.1359 | No |
| 27 | Smtn | 4188 | 0.148 | 0.1264 | No |
| 28 | Schip1 | 4206 | 0.146 | 0.1308 | No |
| 29 | Lsp1 | 4259 | 0.143 | 0.1322 | No |
| 30 | Tpm2 | 4415 | 0.130 | 0.1247 | No |
| 31 | Wwtr1 | 4579 | 0.119 | 0.1161 | No |
| 32 | Lpin1 | 4775 | 0.105 | 0.1044 | No |
| 33 | Mef2a | 5085 | 0.084 | 0.0825 | No |
| 34 | Pygm | 5119 | 0.082 | 0.0831 | No |
| 35 | Mef2c | 5155 | 0.080 | 0.0834 | No |
| 36 | Sptan1 | 5249 | 0.073 | 0.0787 | No |
| 37 | Mapk12 | 5355 | 0.066 | 0.0727 | No |
| 38 | Nav2 | 5480 | 0.057 | 0.0649 | No |
| 39 | Acsl1 | 5495 | 0.056 | 0.0660 | No |
| 40 | Fhl1 | 5505 | 0.055 | 0.0674 | No |
| 41 | Prnp | 5742 | 0.039 | 0.0497 | No |
| 42 | Dtna | 6183 | 0.008 | 0.0142 | No |
| 43 | Agl | 6333 | -0.002 | 0.0021 | No |
| 44 | Kcnh2 | 6494 | -0.012 | -0.0105 | No |
| 45 | Gabarapl2 | 6555 | -0.016 | -0.0147 | No |
| 46 | Ache | 6609 | -0.020 | -0.0182 | No |
| 47 | Tsc2 | 6762 | -0.030 | -0.0295 | No |
| 48 | Svil | 7334 | -0.069 | -0.0732 | No |
| 49 | Tgfb1 | 7357 | -0.071 | -0.0722 | No |
| 50 | Erbb3 | 7524 | -0.081 | -0.0826 | No |
| 51 | Tpm3 | 7678 | -0.094 | -0.0913 | No |
| 52 | Adcy9 | 7782 | -0.101 | -0.0957 | No |
| 53 | Syngr2 | 7866 | -0.107 | -0.0983 | No |
| 54 | Clu | 7877 | -0.108 | -0.0948 | No |
| 55 | Psen2 | 7966 | -0.112 | -0.0975 | No |
| 56 | Camk2b | 7978 | -0.114 | -0.0939 | No |
| 57 | Eif4a2 | 8031 | -0.117 | -0.0936 | No |
| 58 | Gadd45b | 8218 | -0.129 | -0.1036 | No |
| 59 | Mapre3 | 8330 | -0.137 | -0.1072 | No |
| 60 | Chrnb1 | 8355 | -0.140 | -0.1036 | No |
| 61 | Speg | 8403 | -0.143 | -0.1018 | No |
| 62 | Ablim1 | 8410 | -0.143 | -0.0966 | No |
| 63 | Sspn | 8487 | -0.148 | -0.0970 | No |
| 64 | Hdac5 | 8564 | -0.152 | -0.0971 | No |
| 65 | Agrn | 8617 | -0.156 | -0.0952 | No |
| 66 | Sirt2 | 8822 | -0.172 | -0.1050 | No |
| 67 | Rb1 | 8871 | -0.176 | -0.1019 | No |
| 68 | Ak1 | 9033 | -0.187 | -0.1076 | No |
| 69 | Bag1 | 9074 | -0.191 | -0.1034 | No |
| 70 | Tpd52l1 | 9232 | -0.202 | -0.1082 | No |
| 71 | Ryr1 | 9234 | -0.202 | -0.1003 | No |
| 72 | Dmd | 9256 | -0.203 | -0.0939 | No |
| 73 | Ctf1 | 9386 | -0.214 | -0.0960 | No |
| 74 | Ncam1 | 9742 | -0.244 | -0.1152 | No |
| 75 | Flii | 9798 | -0.247 | -0.1099 | No |
| 76 | Plxnb2 | 9877 | -0.254 | -0.1063 | No |
| 77 | Hbegf | 9908 | -0.256 | -0.0986 | No |
| 78 | Mras | 10359 | -0.296 | -0.1235 | No |
| 79 | Pick1 | 10460 | -0.306 | -0.1195 | No |
| 80 | Ifrd1 | 10539 | -0.314 | -0.1134 | No |
| 81 | Igf1 | 10556 | -0.316 | -0.1022 | No |
| 82 | Pde4dip | 10589 | -0.321 | -0.0921 | No |
| 83 | Gnao1 | 10871 | -0.354 | -0.1010 | No |
| 84 | Myo1c | 11593 | -0.465 | -0.1414 | No |
| 85 | Bhlhe40 | 11637 | -0.472 | -0.1262 | No |
| 86 | Large1 | 12109 | -0.635 | -0.1394 | No |
| 87 | Crat | 12149 | -0.667 | -0.1162 | No |
| 88 | Atp6ap1 | 12199 | -0.712 | -0.0920 | No |
| 89 | Col4a2 | 12217 | -0.724 | -0.0648 | No |
| 90 | Sorbs1 | 12307 | -0.872 | -0.0375 | No |
| 91 | Pkia | 12345 | -1.060 | 0.0015 | No |