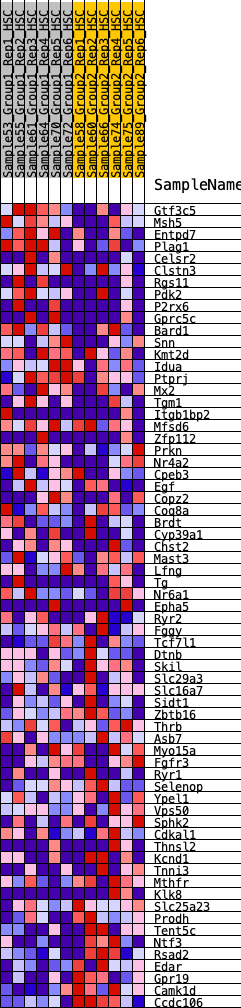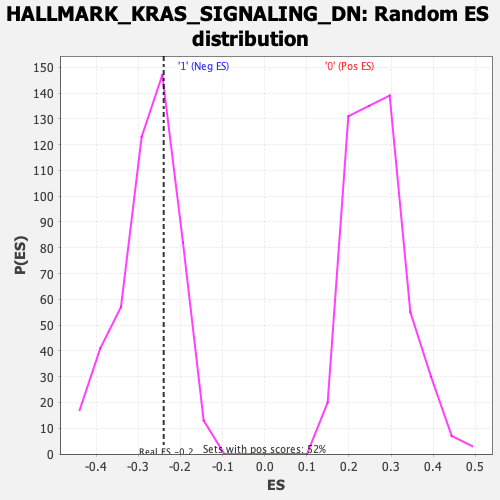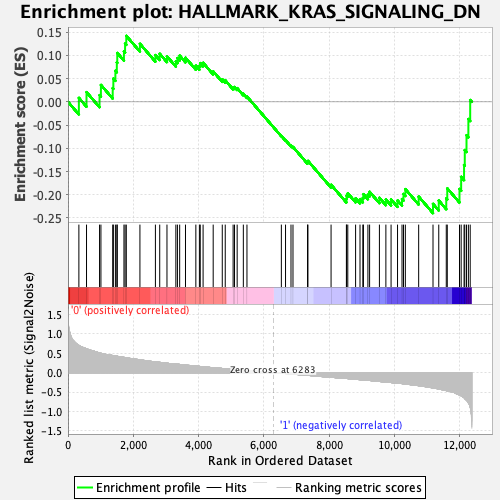
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group2.HSC_Pheno.cls#Group1_versus_Group2_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | -0.23975068 |
| Normalized Enrichment Score (NES) | -0.86689407 |
| Nominal p-value | 0.6666667 |
| FDR q-value | 0.90906525 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gtf3c5 | 333 | 0.703 | 0.0086 | No |
| 2 | Msh5 | 567 | 0.617 | 0.0209 | No |
| 3 | Entpd7 | 966 | 0.510 | 0.0144 | No |
| 4 | Plag1 | 1008 | 0.500 | 0.0364 | No |
| 5 | Celsr2 | 1369 | 0.442 | 0.0295 | No |
| 6 | Clstn3 | 1389 | 0.438 | 0.0502 | No |
| 7 | Rgs11 | 1451 | 0.428 | 0.0670 | No |
| 8 | Pdk2 | 1495 | 0.424 | 0.0849 | No |
| 9 | P2rx6 | 1509 | 0.423 | 0.1053 | No |
| 10 | Gprc5c | 1713 | 0.394 | 0.1088 | No |
| 11 | Bard1 | 1748 | 0.388 | 0.1257 | No |
| 12 | Snn | 1785 | 0.382 | 0.1422 | No |
| 13 | Kmt2d | 2202 | 0.331 | 0.1251 | No |
| 14 | Idua | 2676 | 0.278 | 0.1008 | No |
| 15 | Ptprj | 2808 | 0.266 | 0.1036 | No |
| 16 | Mx2 | 3030 | 0.244 | 0.0980 | No |
| 17 | Tgm1 | 3302 | 0.221 | 0.0872 | No |
| 18 | Itgb1bp2 | 3350 | 0.217 | 0.0944 | No |
| 19 | Mfsd6 | 3419 | 0.212 | 0.0996 | No |
| 20 | Zfp112 | 3597 | 0.195 | 0.0951 | No |
| 21 | Prkn | 3914 | 0.171 | 0.0780 | No |
| 22 | Nr4a2 | 4023 | 0.162 | 0.0774 | No |
| 23 | Cpeb3 | 4049 | 0.159 | 0.0835 | No |
| 24 | Egf | 4136 | 0.152 | 0.0842 | No |
| 25 | Copz2 | 4445 | 0.128 | 0.0656 | No |
| 26 | Coq8a | 4723 | 0.108 | 0.0486 | No |
| 27 | Brdt | 4814 | 0.102 | 0.0464 | No |
| 28 | Cyp39a1 | 5055 | 0.086 | 0.0312 | No |
| 29 | Chst2 | 5098 | 0.083 | 0.0320 | No |
| 30 | Mast3 | 5179 | 0.077 | 0.0295 | No |
| 31 | Lfng | 5368 | 0.065 | 0.0175 | No |
| 32 | Tg | 5479 | 0.057 | 0.0114 | No |
| 33 | Nr6a1 | 6532 | -0.015 | -0.0734 | No |
| 34 | Epha5 | 6660 | -0.023 | -0.0826 | No |
| 35 | Ryr2 | 6827 | -0.035 | -0.0943 | No |
| 36 | Fggy | 6888 | -0.040 | -0.0972 | No |
| 37 | Tcf7l1 | 7333 | -0.069 | -0.1298 | No |
| 38 | Dtnb | 7346 | -0.070 | -0.1272 | No |
| 39 | Skil | 8054 | -0.118 | -0.1787 | No |
| 40 | Slc29a3 | 8523 | -0.150 | -0.2091 | No |
| 41 | Slc16a7 | 8531 | -0.151 | -0.2021 | No |
| 42 | Sidt1 | 8566 | -0.152 | -0.1971 | No |
| 43 | Zbtb16 | 8805 | -0.171 | -0.2078 | No |
| 44 | Thrb | 8940 | -0.180 | -0.2096 | No |
| 45 | Asb7 | 9027 | -0.187 | -0.2071 | No |
| 46 | Myo15a | 9045 | -0.189 | -0.1989 | No |
| 47 | Fgfr3 | 9182 | -0.198 | -0.1999 | No |
| 48 | Ryr1 | 9234 | -0.202 | -0.1938 | No |
| 49 | Selenop | 9534 | -0.227 | -0.2066 | No |
| 50 | Ypel1 | 9732 | -0.243 | -0.2103 | No |
| 51 | Vps50 | 9891 | -0.255 | -0.2102 | No |
| 52 | Sphk2 | 10089 | -0.270 | -0.2126 | No |
| 53 | Cdkal1 | 10226 | -0.282 | -0.2093 | No |
| 54 | Thnsl2 | 10272 | -0.287 | -0.1984 | No |
| 55 | Kcnd1 | 10328 | -0.293 | -0.1881 | No |
| 56 | Tnni3 | 10736 | -0.336 | -0.2041 | No |
| 57 | Mthfr | 11175 | -0.393 | -0.2198 | Yes |
| 58 | Klk8 | 11351 | -0.422 | -0.2127 | Yes |
| 59 | Slc25a23 | 11581 | -0.463 | -0.2078 | Yes |
| 60 | Prodh | 11612 | -0.469 | -0.1865 | Yes |
| 61 | Tent5c | 11986 | -0.575 | -0.1877 | Yes |
| 62 | Ntf3 | 12037 | -0.596 | -0.1615 | Yes |
| 63 | Rsad2 | 12126 | -0.645 | -0.1360 | Yes |
| 64 | Edar | 12148 | -0.667 | -0.1038 | Yes |
| 65 | Gpr19 | 12201 | -0.713 | -0.0719 | Yes |
| 66 | Camk1d | 12258 | -0.779 | -0.0370 | Yes |
| 67 | Ccdc106 | 12314 | -0.896 | 0.0040 | Yes |