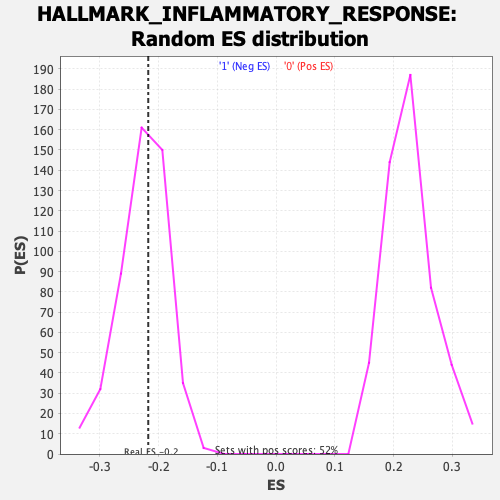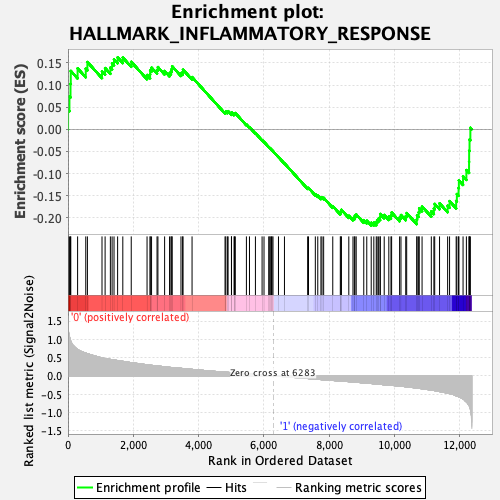
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group2.HSC_Pheno.cls#Group1_versus_Group2_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_INFLAMMATORY_RESPONSE |
| Enrichment Score (ES) | -0.2175354 |
| Normalized Enrichment Score (NES) | -0.9654786 |
| Nominal p-value | 0.51345754 |
| FDR q-value | 0.8437466 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Bst2 | 3 | 1.414 | 0.0442 | No |
| 2 | Gnai3 | 44 | 1.069 | 0.0744 | No |
| 3 | Slc11a2 | 77 | 0.986 | 0.1028 | No |
| 4 | Raf1 | 85 | 0.950 | 0.1320 | No |
| 5 | Csf1 | 294 | 0.726 | 0.1379 | No |
| 6 | Lpar1 | 544 | 0.623 | 0.1371 | No |
| 7 | Gna15 | 594 | 0.606 | 0.1522 | No |
| 8 | Cx3cl1 | 1041 | 0.492 | 0.1312 | No |
| 9 | Slc28a2 | 1136 | 0.475 | 0.1385 | No |
| 10 | Calcrl | 1297 | 0.452 | 0.1396 | No |
| 11 | Tacr1 | 1350 | 0.445 | 0.1494 | No |
| 12 | Rtp4 | 1409 | 0.435 | 0.1583 | No |
| 13 | Rnf144b | 1519 | 0.422 | 0.1626 | No |
| 14 | Fzd5 | 1678 | 0.398 | 0.1622 | No |
| 15 | Ahr | 1936 | 0.362 | 0.1526 | No |
| 16 | Csf3r | 2420 | 0.306 | 0.1229 | No |
| 17 | Lck | 2513 | 0.294 | 0.1246 | No |
| 18 | Pde4b | 2517 | 0.294 | 0.1336 | No |
| 19 | Ifngr2 | 2558 | 0.289 | 0.1394 | No |
| 20 | Slc31a1 | 2726 | 0.273 | 0.1343 | No |
| 21 | Aqp9 | 2753 | 0.270 | 0.1407 | No |
| 22 | Nod2 | 2958 | 0.251 | 0.1319 | No |
| 23 | Mmp14 | 3109 | 0.235 | 0.1271 | No |
| 24 | P2rx7 | 3151 | 0.232 | 0.1310 | No |
| 25 | Icam1 | 3175 | 0.230 | 0.1364 | No |
| 26 | Pcdh7 | 3189 | 0.230 | 0.1425 | No |
| 27 | Irf1 | 3459 | 0.209 | 0.1271 | No |
| 28 | Ccrl2 | 3509 | 0.204 | 0.1295 | No |
| 29 | Il1r1 | 3521 | 0.203 | 0.1350 | No |
| 30 | Dcbld2 | 3798 | 0.179 | 0.1181 | No |
| 31 | Itga5 | 4808 | 0.103 | 0.0390 | No |
| 32 | Tapbp | 4831 | 0.101 | 0.0404 | No |
| 33 | Gpc3 | 4880 | 0.099 | 0.0396 | No |
| 34 | Icosl | 4901 | 0.098 | 0.0410 | No |
| 35 | Ldlr | 5004 | 0.090 | 0.0355 | No |
| 36 | Scarf1 | 5006 | 0.090 | 0.0383 | No |
| 37 | Acvr2a | 5084 | 0.084 | 0.0346 | No |
| 38 | Chst2 | 5098 | 0.083 | 0.0362 | No |
| 39 | Ifnar1 | 5122 | 0.082 | 0.0369 | No |
| 40 | Met | 5462 | 0.058 | 0.0110 | No |
| 41 | Selenos | 5556 | 0.052 | 0.0051 | No |
| 42 | Tlr2 | 5738 | 0.039 | -0.0085 | No |
| 43 | Nfkbia | 5942 | 0.023 | -0.0243 | No |
| 44 | Gabbr1 | 6001 | 0.020 | -0.0284 | No |
| 45 | Rela | 6145 | 0.010 | -0.0398 | No |
| 46 | Rgs1 | 6178 | 0.008 | -0.0421 | No |
| 47 | Slc7a1 | 6191 | 0.007 | -0.0429 | No |
| 48 | Sell | 6231 | 0.004 | -0.0459 | No |
| 49 | Axl | 6246 | 0.003 | -0.0470 | No |
| 50 | Ripk2 | 6276 | 0.001 | -0.0493 | No |
| 51 | Ptafr | 6444 | -0.009 | -0.0627 | No |
| 52 | Irf7 | 6627 | -0.021 | -0.0769 | No |
| 53 | Nmi | 7337 | -0.070 | -0.1325 | No |
| 54 | Myc | 7363 | -0.071 | -0.1323 | No |
| 55 | Gpr132 | 7572 | -0.085 | -0.1467 | No |
| 56 | Atp2c1 | 7647 | -0.091 | -0.1498 | No |
| 57 | Il15ra | 7744 | -0.098 | -0.1546 | No |
| 58 | Sri | 7776 | -0.101 | -0.1540 | No |
| 59 | Eif2ak2 | 7827 | -0.104 | -0.1548 | No |
| 60 | Tnfrsf1b | 8106 | -0.122 | -0.1736 | No |
| 61 | Rhog | 8337 | -0.138 | -0.1881 | No |
| 62 | Il18 | 8354 | -0.140 | -0.1850 | No |
| 63 | Ebi3 | 8371 | -0.141 | -0.1818 | No |
| 64 | Osmr | 8597 | -0.155 | -0.1953 | No |
| 65 | Ptger4 | 8725 | -0.164 | -0.2006 | No |
| 66 | Slc31a2 | 8767 | -0.167 | -0.1987 | No |
| 67 | Lyn | 8785 | -0.169 | -0.1948 | No |
| 68 | Gpr183 | 8823 | -0.172 | -0.1924 | No |
| 69 | Lcp2 | 9055 | -0.190 | -0.2052 | No |
| 70 | Hif1a | 9147 | -0.196 | -0.2065 | No |
| 71 | Klf6 | 9283 | -0.206 | -0.2111 | Yes |
| 72 | Pik3r5 | 9363 | -0.213 | -0.2109 | Yes |
| 73 | Abi1 | 9437 | -0.219 | -0.2099 | Yes |
| 74 | Atp2b1 | 9464 | -0.221 | -0.2051 | Yes |
| 75 | Inhba | 9510 | -0.225 | -0.2017 | Yes |
| 76 | Tlr3 | 9556 | -0.229 | -0.1982 | Yes |
| 77 | Plaur | 9560 | -0.229 | -0.1913 | Yes |
| 78 | Tnfsf10 | 9681 | -0.240 | -0.1935 | Yes |
| 79 | Adrm1 | 9822 | -0.249 | -0.1971 | Yes |
| 80 | Il18rap | 9888 | -0.254 | -0.1944 | Yes |
| 81 | Hbegf | 9908 | -0.256 | -0.1879 | Yes |
| 82 | P2ry2 | 10152 | -0.276 | -0.1991 | Yes |
| 83 | Psen1 | 10194 | -0.280 | -0.1937 | Yes |
| 84 | Sema4d | 10340 | -0.294 | -0.1963 | Yes |
| 85 | Il18r1 | 10369 | -0.296 | -0.1893 | Yes |
| 86 | Pvr | 10677 | -0.329 | -0.2040 | Yes |
| 87 | Ptpre | 10687 | -0.331 | -0.1943 | Yes |
| 88 | Irak2 | 10735 | -0.336 | -0.1876 | Yes |
| 89 | Nfkb1 | 10753 | -0.339 | -0.1784 | Yes |
| 90 | Nampt | 10840 | -0.350 | -0.1744 | Yes |
| 91 | Cd82 | 11123 | -0.386 | -0.1853 | Yes |
| 92 | Il10ra | 11202 | -0.396 | -0.1792 | Yes |
| 93 | Itgb3 | 11227 | -0.400 | -0.1686 | Yes |
| 94 | P2rx4 | 11375 | -0.426 | -0.1672 | Yes |
| 95 | Il4ra | 11623 | -0.470 | -0.1726 | Yes |
| 96 | Emp3 | 11682 | -0.482 | -0.1622 | Yes |
| 97 | Kif1b | 11884 | -0.541 | -0.1616 | Yes |
| 98 | Acvr1b | 11906 | -0.548 | -0.1462 | Yes |
| 99 | Abca1 | 11958 | -0.565 | -0.1326 | Yes |
| 100 | Ly6e | 11967 | -0.569 | -0.1154 | Yes |
| 101 | Gch1 | 12095 | -0.626 | -0.1061 | Yes |
| 102 | Ifitm1 | 12197 | -0.710 | -0.0920 | Yes |
| 103 | Atp2a2 | 12279 | -0.804 | -0.0734 | Yes |
| 104 | Mxd1 | 12282 | -0.809 | -0.0482 | Yes |
| 105 | Slamf1 | 12294 | -0.832 | -0.0230 | Yes |
| 106 | Il15 | 12319 | -0.908 | 0.0036 | Yes |