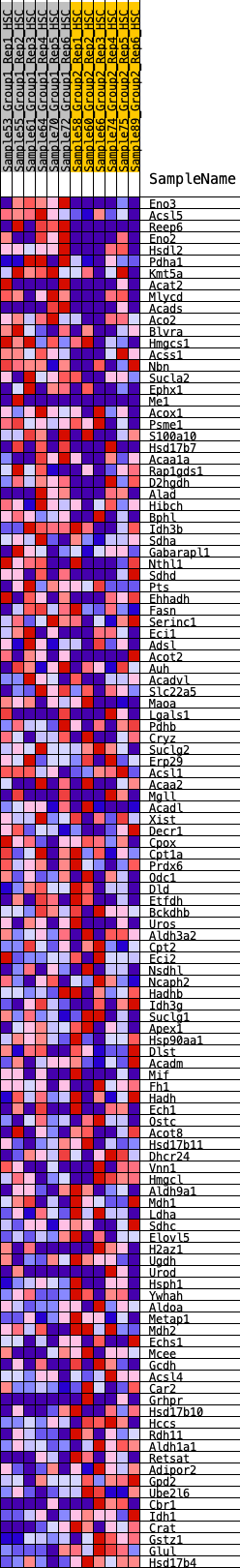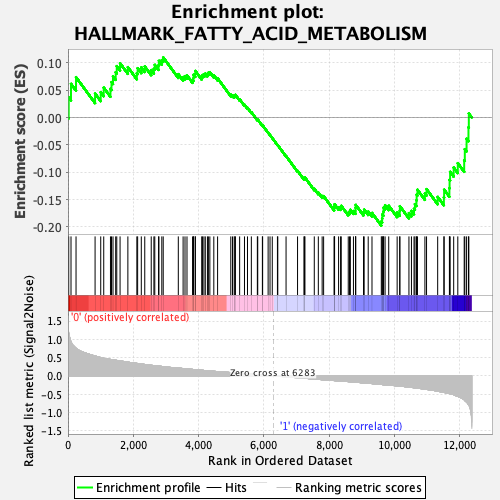
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group2.HSC_Pheno.cls#Group1_versus_Group2_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
| Enrichment Score (ES) | -0.19755685 |
| Normalized Enrichment Score (NES) | -0.837367 |
| Nominal p-value | 0.8038835 |
| FDR q-value | 0.8858759 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Eno3 | 22 | 1.180 | 0.0367 | No |
| 2 | Acsl5 | 92 | 0.937 | 0.0617 | No |
| 3 | Reep6 | 246 | 0.756 | 0.0739 | No |
| 4 | Eno2 | 829 | 0.542 | 0.0441 | No |
| 5 | Hsdl2 | 1002 | 0.503 | 0.0464 | No |
| 6 | Pdha1 | 1094 | 0.483 | 0.0548 | No |
| 7 | Kmt5a | 1300 | 0.450 | 0.0527 | No |
| 8 | Acat2 | 1332 | 0.447 | 0.0648 | No |
| 9 | Mlycd | 1378 | 0.441 | 0.0755 | No |
| 10 | Acads | 1460 | 0.428 | 0.0829 | No |
| 11 | Aco2 | 1493 | 0.424 | 0.0941 | No |
| 12 | Blvra | 1594 | 0.410 | 0.0994 | No |
| 13 | Hmgcs1 | 1833 | 0.376 | 0.0922 | No |
| 14 | Acss1 | 2108 | 0.343 | 0.0811 | No |
| 15 | Nbn | 2132 | 0.340 | 0.0903 | No |
| 16 | Sucla2 | 2244 | 0.325 | 0.0918 | No |
| 17 | Ephx1 | 2350 | 0.314 | 0.0935 | No |
| 18 | Me1 | 2548 | 0.290 | 0.0869 | No |
| 19 | Acox1 | 2630 | 0.282 | 0.0895 | No |
| 20 | Psme1 | 2657 | 0.279 | 0.0965 | No |
| 21 | S100a10 | 2773 | 0.269 | 0.0959 | No |
| 22 | Hsd17b7 | 2783 | 0.268 | 0.1039 | No |
| 23 | Acaa1a | 2872 | 0.259 | 0.1052 | No |
| 24 | Rap1gds1 | 2915 | 0.255 | 0.1100 | No |
| 25 | D2hgdh | 3377 | 0.216 | 0.0794 | No |
| 26 | Alad | 3524 | 0.203 | 0.0741 | No |
| 27 | Hibch | 3582 | 0.196 | 0.0759 | No |
| 28 | Bphl | 3643 | 0.192 | 0.0773 | No |
| 29 | Idh3b | 3813 | 0.179 | 0.0693 | No |
| 30 | Sdha | 3843 | 0.176 | 0.0727 | No |
| 31 | Gabarapl1 | 3845 | 0.176 | 0.0784 | No |
| 32 | Nthl1 | 3896 | 0.172 | 0.0799 | No |
| 33 | Sdhd | 3899 | 0.172 | 0.0853 | No |
| 34 | Pts | 4094 | 0.155 | 0.0746 | No |
| 35 | Ehhadh | 4116 | 0.153 | 0.0779 | No |
| 36 | Fasn | 4161 | 0.150 | 0.0792 | No |
| 37 | Serinc1 | 4203 | 0.146 | 0.0806 | No |
| 38 | Eci1 | 4269 | 0.142 | 0.0799 | No |
| 39 | Adsl | 4298 | 0.139 | 0.0822 | No |
| 40 | Acot2 | 4343 | 0.135 | 0.0830 | No |
| 41 | Auh | 4464 | 0.127 | 0.0774 | No |
| 42 | Acadvl | 4580 | 0.118 | 0.0718 | No |
| 43 | Slc22a5 | 4984 | 0.092 | 0.0419 | No |
| 44 | Maoa | 5037 | 0.087 | 0.0405 | No |
| 45 | Lgals1 | 5088 | 0.084 | 0.0392 | No |
| 46 | Pdhb | 5110 | 0.082 | 0.0401 | No |
| 47 | Cryz | 5125 | 0.082 | 0.0416 | No |
| 48 | Suclg2 | 5255 | 0.072 | 0.0335 | No |
| 49 | Erp29 | 5403 | 0.061 | 0.0235 | No |
| 50 | Acsl1 | 5495 | 0.056 | 0.0179 | No |
| 51 | Acaa2 | 5613 | 0.048 | 0.0099 | No |
| 52 | Mgll | 5797 | 0.036 | -0.0039 | No |
| 53 | Acadl | 5809 | 0.035 | -0.0036 | No |
| 54 | Xist | 5951 | 0.023 | -0.0144 | No |
| 55 | Decr1 | 5959 | 0.022 | -0.0143 | No |
| 56 | Cpox | 6127 | 0.011 | -0.0275 | No |
| 57 | Cpt1a | 6185 | 0.007 | -0.0320 | No |
| 58 | Prdx6 | 6254 | 0.002 | -0.0374 | No |
| 59 | Odc1 | 6412 | -0.008 | -0.0500 | No |
| 60 | Dld | 6423 | -0.008 | -0.0505 | No |
| 61 | Etfdh | 6676 | -0.023 | -0.0704 | No |
| 62 | Bckdhb | 7028 | -0.048 | -0.0975 | No |
| 63 | Uros | 7224 | -0.061 | -0.1114 | No |
| 64 | Aldh3a2 | 7239 | -0.062 | -0.1105 | No |
| 65 | Cpt2 | 7254 | -0.063 | -0.1096 | No |
| 66 | Eci2 | 7540 | -0.082 | -0.1302 | No |
| 67 | Nsdhl | 7664 | -0.092 | -0.1372 | No |
| 68 | Ncaph2 | 7781 | -0.101 | -0.1434 | No |
| 69 | Hadhb | 7826 | -0.103 | -0.1436 | No |
| 70 | Idh3g | 8148 | -0.125 | -0.1658 | No |
| 71 | Suclg1 | 8155 | -0.126 | -0.1621 | No |
| 72 | Apex1 | 8162 | -0.126 | -0.1585 | No |
| 73 | Hsp90aa1 | 8284 | -0.134 | -0.1640 | No |
| 74 | Dlst | 8345 | -0.139 | -0.1644 | No |
| 75 | Acadm | 8365 | -0.141 | -0.1613 | No |
| 76 | Mif | 8580 | -0.153 | -0.1738 | No |
| 77 | Fh1 | 8619 | -0.156 | -0.1718 | No |
| 78 | Hadh | 8644 | -0.158 | -0.1686 | No |
| 79 | Ech1 | 8739 | -0.165 | -0.1709 | No |
| 80 | Ostc | 8803 | -0.170 | -0.1705 | No |
| 81 | Acot8 | 8804 | -0.171 | -0.1649 | No |
| 82 | Hsd17b11 | 8809 | -0.171 | -0.1597 | No |
| 83 | Dhcr24 | 9047 | -0.189 | -0.1729 | No |
| 84 | Vnn1 | 9067 | -0.190 | -0.1682 | No |
| 85 | Hmgcl | 9191 | -0.199 | -0.1718 | No |
| 86 | Aldh9a1 | 9307 | -0.208 | -0.1744 | No |
| 87 | Mdh1 | 9592 | -0.232 | -0.1900 | Yes |
| 88 | Ldha | 9618 | -0.234 | -0.1844 | Yes |
| 89 | Sdhc | 9623 | -0.234 | -0.1771 | Yes |
| 90 | Elovl5 | 9656 | -0.237 | -0.1719 | Yes |
| 91 | H2az1 | 9663 | -0.238 | -0.1646 | Yes |
| 92 | Ugdh | 9708 | -0.242 | -0.1603 | Yes |
| 93 | Urod | 9819 | -0.249 | -0.1612 | Yes |
| 94 | Hsph1 | 10077 | -0.269 | -0.1734 | Yes |
| 95 | Ywhah | 10156 | -0.276 | -0.1707 | Yes |
| 96 | Aldoa | 10161 | -0.277 | -0.1620 | Yes |
| 97 | Metap1 | 10438 | -0.305 | -0.1746 | Yes |
| 98 | Mdh2 | 10515 | -0.312 | -0.1706 | Yes |
| 99 | Echs1 | 10594 | -0.321 | -0.1665 | Yes |
| 100 | Mcee | 10622 | -0.323 | -0.1582 | Yes |
| 101 | Gcdh | 10668 | -0.327 | -0.1512 | Yes |
| 102 | Acsl4 | 10675 | -0.329 | -0.1409 | Yes |
| 103 | Car2 | 10699 | -0.331 | -0.1320 | Yes |
| 104 | Grhpr | 10927 | -0.359 | -0.1388 | Yes |
| 105 | Hsd17b10 | 10977 | -0.365 | -0.1309 | Yes |
| 106 | Hccs | 11318 | -0.415 | -0.1451 | Yes |
| 107 | Rdh11 | 11508 | -0.448 | -0.1459 | Yes |
| 108 | Aldh1a1 | 11516 | -0.450 | -0.1317 | Yes |
| 109 | Retsat | 11677 | -0.480 | -0.1291 | Yes |
| 110 | Adipor2 | 11683 | -0.482 | -0.1138 | Yes |
| 111 | Gpd2 | 11699 | -0.485 | -0.0992 | Yes |
| 112 | Ube2l6 | 11808 | -0.510 | -0.0914 | Yes |
| 113 | Cbr1 | 11934 | -0.557 | -0.0834 | Yes |
| 114 | Idh1 | 12130 | -0.650 | -0.0781 | Yes |
| 115 | Crat | 12149 | -0.667 | -0.0577 | Yes |
| 116 | Gstz1 | 12203 | -0.716 | -0.0387 | Yes |
| 117 | Glul | 12261 | -0.781 | -0.0178 | Yes |
| 118 | Hsd17b4 | 12272 | -0.798 | 0.0074 | Yes |