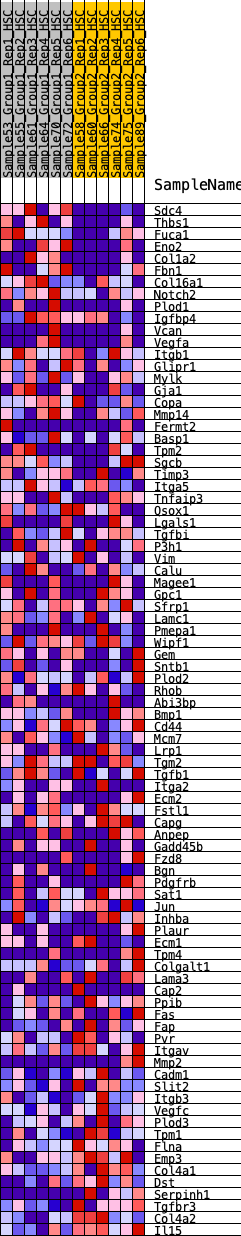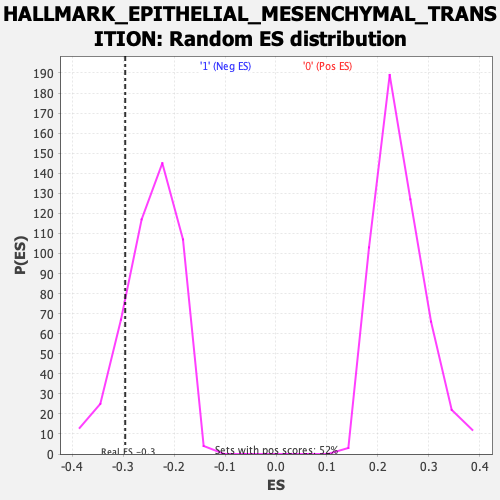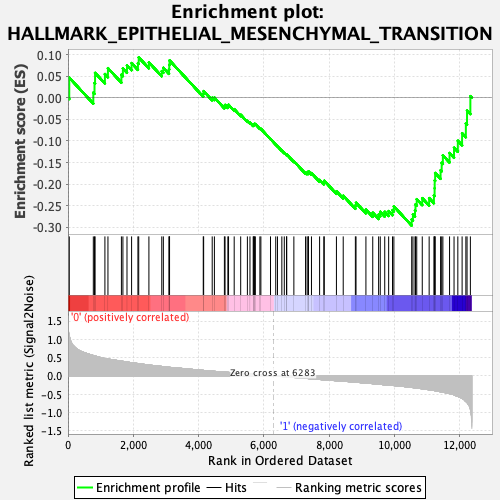
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group2.HSC_Pheno.cls#Group1_versus_Group2_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | -0.29597712 |
| Normalized Enrichment Score (NES) | -1.2012868 |
| Nominal p-value | 0.16945606 |
| FDR q-value | 0.49647897 |
| FWER p-Value | 0.909 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sdc4 | 39 | 1.084 | 0.0463 | No |
| 2 | Thbs1 | 775 | 0.558 | 0.0119 | No |
| 3 | Fuca1 | 812 | 0.549 | 0.0340 | No |
| 4 | Eno2 | 829 | 0.542 | 0.0574 | No |
| 5 | Col1a2 | 1130 | 0.476 | 0.0547 | No |
| 6 | Fbn1 | 1223 | 0.463 | 0.0683 | No |
| 7 | Col16a1 | 1635 | 0.405 | 0.0533 | No |
| 8 | Notch2 | 1681 | 0.398 | 0.0678 | No |
| 9 | Plod1 | 1806 | 0.379 | 0.0751 | No |
| 10 | Igfbp4 | 1947 | 0.361 | 0.0801 | No |
| 11 | Vcan | 2141 | 0.339 | 0.0799 | No |
| 12 | Vegfa | 2165 | 0.335 | 0.0933 | No |
| 13 | Itgb1 | 2477 | 0.299 | 0.0816 | No |
| 14 | Glipr1 | 2871 | 0.259 | 0.0615 | No |
| 15 | Mylk | 2919 | 0.255 | 0.0692 | No |
| 16 | Gja1 | 3092 | 0.237 | 0.0660 | No |
| 17 | Copa | 3101 | 0.236 | 0.0762 | No |
| 18 | Mmp14 | 3109 | 0.235 | 0.0863 | No |
| 19 | Fermt2 | 4141 | 0.152 | 0.0093 | No |
| 20 | Basp1 | 4153 | 0.151 | 0.0153 | No |
| 21 | Tpm2 | 4415 | 0.130 | -0.0001 | No |
| 22 | Sgcb | 4482 | 0.125 | 0.0003 | No |
| 23 | Timp3 | 4788 | 0.104 | -0.0198 | No |
| 24 | Itga5 | 4808 | 0.103 | -0.0167 | No |
| 25 | Tnfaip3 | 4895 | 0.098 | -0.0192 | No |
| 26 | Qsox1 | 4915 | 0.097 | -0.0163 | No |
| 27 | Lgals1 | 5088 | 0.084 | -0.0265 | No |
| 28 | Tgfbi | 5287 | 0.070 | -0.0394 | No |
| 29 | P3h1 | 5492 | 0.056 | -0.0535 | No |
| 30 | Vim | 5571 | 0.051 | -0.0575 | No |
| 31 | Calu | 5670 | 0.044 | -0.0635 | No |
| 32 | Magee1 | 5685 | 0.043 | -0.0626 | No |
| 33 | Gpc1 | 5696 | 0.043 | -0.0615 | No |
| 34 | Sfrp1 | 5708 | 0.042 | -0.0605 | No |
| 35 | Lamc1 | 5736 | 0.040 | -0.0609 | No |
| 36 | Pmepa1 | 5875 | 0.029 | -0.0708 | No |
| 37 | Wipf1 | 5905 | 0.026 | -0.0720 | No |
| 38 | Gem | 6200 | 0.006 | -0.0956 | No |
| 39 | Sntb1 | 6360 | -0.004 | -0.1084 | No |
| 40 | Plod2 | 6413 | -0.008 | -0.1123 | No |
| 41 | Rhob | 6547 | -0.016 | -0.1224 | No |
| 42 | Abi3bp | 6622 | -0.021 | -0.1275 | No |
| 43 | Bmp1 | 6691 | -0.024 | -0.1319 | No |
| 44 | Cd44 | 6696 | -0.025 | -0.1311 | No |
| 45 | Mcm7 | 6915 | -0.041 | -0.1470 | No |
| 46 | Lrp1 | 7273 | -0.064 | -0.1731 | No |
| 47 | Tgm2 | 7321 | -0.068 | -0.1738 | No |
| 48 | Tgfb1 | 7357 | -0.071 | -0.1735 | No |
| 49 | Itga2 | 7361 | -0.071 | -0.1705 | No |
| 50 | Ecm2 | 7454 | -0.077 | -0.1744 | No |
| 51 | Fstl1 | 7703 | -0.095 | -0.1903 | No |
| 52 | Capg | 7833 | -0.104 | -0.1961 | No |
| 53 | Anpep | 7845 | -0.105 | -0.1922 | No |
| 54 | Gadd45b | 8218 | -0.129 | -0.2166 | No |
| 55 | Fzd8 | 8426 | -0.144 | -0.2269 | No |
| 56 | Bgn | 8798 | -0.170 | -0.2493 | No |
| 57 | Pdgfrb | 8818 | -0.172 | -0.2431 | No |
| 58 | Sat1 | 9121 | -0.194 | -0.2588 | No |
| 59 | Jun | 9330 | -0.210 | -0.2662 | No |
| 60 | Inhba | 9510 | -0.225 | -0.2705 | No |
| 61 | Plaur | 9560 | -0.229 | -0.2640 | No |
| 62 | Ecm1 | 9697 | -0.241 | -0.2641 | No |
| 63 | Tpm4 | 9817 | -0.249 | -0.2624 | No |
| 64 | Colgalt1 | 9935 | -0.258 | -0.2602 | No |
| 65 | Lama3 | 9977 | -0.261 | -0.2516 | No |
| 66 | Cap2 | 10523 | -0.313 | -0.2817 | Yes |
| 67 | Ppib | 10561 | -0.317 | -0.2702 | Yes |
| 68 | Fas | 10624 | -0.323 | -0.2606 | Yes |
| 69 | Fap | 10639 | -0.324 | -0.2469 | Yes |
| 70 | Pvr | 10677 | -0.329 | -0.2349 | Yes |
| 71 | Itgav | 10846 | -0.351 | -0.2326 | Yes |
| 72 | Mmp2 | 11057 | -0.378 | -0.2324 | Yes |
| 73 | Cadm1 | 11203 | -0.396 | -0.2262 | Yes |
| 74 | Slit2 | 11224 | -0.399 | -0.2096 | Yes |
| 75 | Itgb3 | 11227 | -0.400 | -0.1915 | Yes |
| 76 | Vegfc | 11241 | -0.402 | -0.1742 | Yes |
| 77 | Plod3 | 11407 | -0.432 | -0.1679 | Yes |
| 78 | Tpm1 | 11441 | -0.438 | -0.1507 | Yes |
| 79 | Flna | 11477 | -0.442 | -0.1333 | Yes |
| 80 | Emp3 | 11682 | -0.482 | -0.1280 | Yes |
| 81 | Col4a1 | 11819 | -0.512 | -0.1157 | Yes |
| 82 | Dst | 11935 | -0.557 | -0.0996 | Yes |
| 83 | Serpinh1 | 12066 | -0.612 | -0.0823 | Yes |
| 84 | Tgfbr3 | 12181 | -0.696 | -0.0598 | Yes |
| 85 | Col4a2 | 12217 | -0.724 | -0.0296 | Yes |
| 86 | Il15 | 12319 | -0.908 | 0.0036 | Yes |