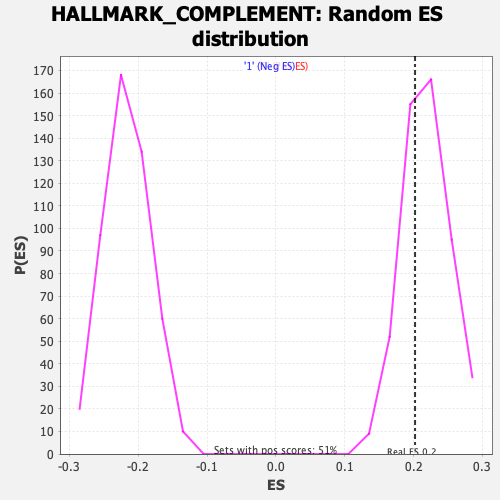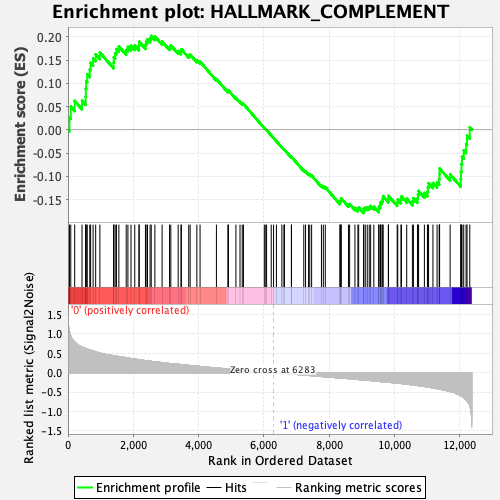
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group2.HSC_Pheno.cls#Group1_versus_Group2_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_COMPLEMENT |
| Enrichment Score (ES) | 0.20226298 |
| Normalized Enrichment Score (NES) | 0.92482895 |
| Nominal p-value | 0.66536206 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gnai3 | 44 | 1.069 | 0.0265 | Yes |
| 2 | Raf1 | 85 | 0.950 | 0.0499 | Yes |
| 3 | Ctsd | 205 | 0.789 | 0.0624 | Yes |
| 4 | Casp4 | 429 | 0.660 | 0.0628 | Yes |
| 5 | Rnf4 | 541 | 0.625 | 0.0713 | Yes |
| 6 | Cpm | 546 | 0.623 | 0.0885 | Yes |
| 7 | Fyn | 558 | 0.619 | 0.1050 | Yes |
| 8 | Ehd1 | 589 | 0.609 | 0.1197 | Yes |
| 9 | F5 | 667 | 0.587 | 0.1299 | Yes |
| 10 | Pfn1 | 693 | 0.581 | 0.1442 | Yes |
| 11 | Lap3 | 769 | 0.560 | 0.1538 | Yes |
| 12 | Dock4 | 847 | 0.539 | 0.1627 | Yes |
| 13 | Stx4a | 976 | 0.509 | 0.1665 | Yes |
| 14 | Akap10 | 1395 | 0.437 | 0.1447 | Yes |
| 15 | Grb2 | 1403 | 0.435 | 0.1563 | Yes |
| 16 | Maff | 1444 | 0.429 | 0.1652 | Yes |
| 17 | Atox1 | 1483 | 0.425 | 0.1740 | Yes |
| 18 | Spock2 | 1557 | 0.415 | 0.1797 | Yes |
| 19 | Pdgfb | 1782 | 0.382 | 0.1722 | Yes |
| 20 | Gng2 | 1834 | 0.376 | 0.1786 | Yes |
| 21 | Usp14 | 1929 | 0.363 | 0.1812 | Yes |
| 22 | Prdm4 | 2044 | 0.349 | 0.1816 | Yes |
| 23 | Was | 2174 | 0.334 | 0.1805 | Yes |
| 24 | Rabif | 2178 | 0.333 | 0.1896 | Yes |
| 25 | F8 | 2371 | 0.311 | 0.1827 | Yes |
| 26 | Ctsh | 2391 | 0.309 | 0.1899 | Yes |
| 27 | Dusp6 | 2434 | 0.305 | 0.1950 | Yes |
| 28 | Lck | 2513 | 0.294 | 0.1969 | Yes |
| 29 | Me1 | 2548 | 0.290 | 0.2023 | Yes |
| 30 | Dgkh | 2660 | 0.279 | 0.2011 | No |
| 31 | Dock10 | 2881 | 0.258 | 0.1903 | No |
| 32 | Mmp14 | 3109 | 0.235 | 0.1784 | No |
| 33 | Plek | 3148 | 0.233 | 0.1819 | No |
| 34 | Jak2 | 3373 | 0.216 | 0.1696 | No |
| 35 | Irf1 | 3459 | 0.209 | 0.1686 | No |
| 36 | Brpf3 | 3471 | 0.207 | 0.1735 | No |
| 37 | Gp9 | 3696 | 0.187 | 0.1605 | No |
| 38 | Dgkg | 3740 | 0.184 | 0.1621 | No |
| 39 | Ctsl | 3945 | 0.168 | 0.1502 | No |
| 40 | Vcpip1 | 4043 | 0.159 | 0.1467 | No |
| 41 | Pla2g4a | 4544 | 0.122 | 0.1093 | No |
| 42 | Tnfaip3 | 4895 | 0.098 | 0.0835 | No |
| 43 | Ctsc | 4912 | 0.097 | 0.0849 | No |
| 44 | Fdx1 | 5141 | 0.081 | 0.0686 | No |
| 45 | Calm1 | 5266 | 0.072 | 0.0605 | No |
| 46 | Cd46 | 5334 | 0.067 | 0.0569 | No |
| 47 | Casp9 | 5372 | 0.064 | 0.0557 | No |
| 48 | Casp7 | 6009 | 0.019 | 0.0043 | No |
| 49 | Sirt6 | 6038 | 0.017 | 0.0025 | No |
| 50 | Calm3 | 6080 | 0.014 | -0.0005 | No |
| 51 | Cr2 | 6222 | 0.005 | -0.0119 | No |
| 52 | S100a9 | 6294 | 0.000 | -0.0177 | No |
| 53 | Adam9 | 6381 | -0.006 | -0.0245 | No |
| 54 | Kif2a | 6548 | -0.016 | -0.0377 | No |
| 55 | Sh2b3 | 6611 | -0.020 | -0.0422 | No |
| 56 | Irf7 | 6627 | -0.021 | -0.0428 | No |
| 57 | Hspa5 | 6839 | -0.036 | -0.0590 | No |
| 58 | Lta4h | 6842 | -0.036 | -0.0582 | No |
| 59 | Lipa | 7219 | -0.061 | -0.0872 | No |
| 60 | Lrp1 | 7273 | -0.064 | -0.0897 | No |
| 61 | Casp1 | 7365 | -0.071 | -0.0951 | No |
| 62 | Cblb | 7402 | -0.073 | -0.0960 | No |
| 63 | Kcnip3 | 7459 | -0.077 | -0.0984 | No |
| 64 | S100a13 | 7759 | -0.099 | -0.1200 | No |
| 65 | Prkcd | 7817 | -0.103 | -0.1218 | No |
| 66 | Clu | 7877 | -0.108 | -0.1236 | No |
| 67 | Msrb1 | 8323 | -0.137 | -0.1561 | No |
| 68 | Rhog | 8337 | -0.138 | -0.1533 | No |
| 69 | Gnb2 | 8356 | -0.140 | -0.1508 | No |
| 70 | Cpq | 8364 | -0.140 | -0.1474 | No |
| 71 | Usp8 | 8587 | -0.154 | -0.1612 | No |
| 72 | Dpp4 | 8622 | -0.157 | -0.1596 | No |
| 73 | Lyn | 8785 | -0.169 | -0.1681 | No |
| 74 | Xpnpep1 | 8876 | -0.176 | -0.1705 | No |
| 75 | Dusp5 | 8899 | -0.178 | -0.1673 | No |
| 76 | Timp2 | 9048 | -0.189 | -0.1740 | No |
| 77 | Lcp2 | 9055 | -0.190 | -0.1692 | No |
| 78 | Dyrk2 | 9101 | -0.192 | -0.1675 | No |
| 79 | Gnai2 | 9162 | -0.197 | -0.1668 | No |
| 80 | Ctsb | 9222 | -0.201 | -0.1660 | No |
| 81 | Ctss | 9266 | -0.204 | -0.1638 | No |
| 82 | Pik3r5 | 9363 | -0.213 | -0.1656 | No |
| 83 | Pdp1 | 9511 | -0.225 | -0.1713 | No |
| 84 | Src | 9519 | -0.226 | -0.1655 | No |
| 85 | Plaur | 9560 | -0.229 | -0.1624 | No |
| 86 | Usp16 | 9567 | -0.229 | -0.1564 | No |
| 87 | Lamp2 | 9611 | -0.233 | -0.1533 | No |
| 88 | Gmfb | 9637 | -0.236 | -0.1488 | No |
| 89 | Prep | 9646 | -0.236 | -0.1428 | No |
| 90 | Rbsn | 9809 | -0.248 | -0.1490 | No |
| 91 | Rce1 | 9811 | -0.248 | -0.1421 | No |
| 92 | Pik3cg | 10078 | -0.269 | -0.1563 | No |
| 93 | Zeb1 | 10096 | -0.270 | -0.1501 | No |
| 94 | Psen1 | 10194 | -0.280 | -0.1501 | No |
| 95 | Gnb4 | 10208 | -0.280 | -0.1433 | No |
| 96 | Kynu | 10370 | -0.296 | -0.1481 | No |
| 97 | Psmb9 | 10544 | -0.315 | -0.1534 | No |
| 98 | Pclo | 10575 | -0.319 | -0.1469 | No |
| 99 | Car2 | 10699 | -0.331 | -0.1476 | No |
| 100 | Ppp2cb | 10714 | -0.333 | -0.1394 | No |
| 101 | Itgam | 10734 | -0.336 | -0.1315 | No |
| 102 | Pim1 | 10911 | -0.358 | -0.1358 | No |
| 103 | Ctso | 11004 | -0.369 | -0.1329 | No |
| 104 | Ppp4c | 11019 | -0.372 | -0.1236 | No |
| 105 | Gca | 11039 | -0.375 | -0.1146 | No |
| 106 | Prss36 | 11173 | -0.393 | -0.1144 | No |
| 107 | Gata3 | 11300 | -0.412 | -0.1131 | No |
| 108 | Csrp1 | 11364 | -0.424 | -0.1063 | No |
| 109 | Irf2 | 11374 | -0.426 | -0.0951 | No |
| 110 | Usp15 | 11377 | -0.426 | -0.0833 | No |
| 111 | Gpd2 | 11699 | -0.485 | -0.0959 | No |
| 112 | Casp3 | 12023 | -0.589 | -0.1057 | No |
| 113 | Plscr1 | 12033 | -0.594 | -0.0897 | No |
| 114 | Dock9 | 12047 | -0.600 | -0.0739 | No |
| 115 | Anxa5 | 12063 | -0.609 | -0.0580 | No |
| 116 | Pik3ca | 12115 | -0.639 | -0.0442 | No |
| 117 | C2 | 12188 | -0.701 | -0.0303 | No |
| 118 | Col4a2 | 12217 | -0.724 | -0.0123 | No |
| 119 | Prcp | 12301 | -0.857 | 0.0051 | No |