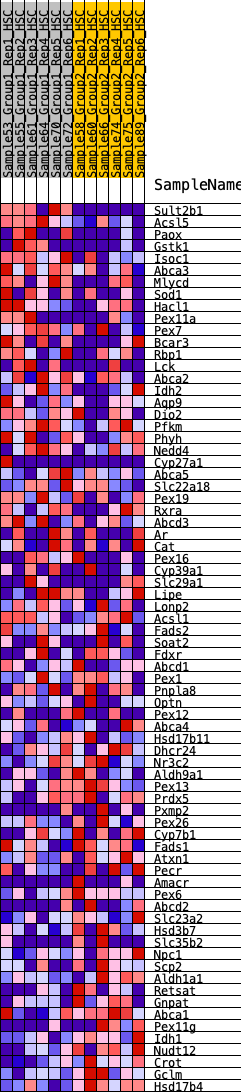
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group2.HSC_Pheno.cls#Group1_versus_Group2_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.2811895 |
| Normalized Enrichment Score (NES) | -1.2816557 |
| Nominal p-value | 0.056842104 |
| FDR q-value | 0.4018878 |
| FWER p-Value | 0.778 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sult2b1 | 59 | 1.032 | 0.0380 | No |
| 2 | Acsl5 | 92 | 0.937 | 0.0742 | No |
| 3 | Paox | 364 | 0.685 | 0.0806 | No |
| 4 | Gstk1 | 393 | 0.674 | 0.1063 | No |
| 5 | Isoc1 | 438 | 0.658 | 0.1300 | No |
| 6 | Abca3 | 514 | 0.632 | 0.1501 | No |
| 7 | Mlycd | 1378 | 0.441 | 0.0982 | No |
| 8 | Sod1 | 1670 | 0.399 | 0.0910 | No |
| 9 | Hacl1 | 1707 | 0.395 | 0.1045 | No |
| 10 | Pex11a | 2127 | 0.341 | 0.0845 | No |
| 11 | Pex7 | 2152 | 0.337 | 0.0965 | No |
| 12 | Bcar3 | 2232 | 0.327 | 0.1037 | No |
| 13 | Rbp1 | 2234 | 0.326 | 0.1171 | No |
| 14 | Lck | 2513 | 0.294 | 0.1067 | No |
| 15 | Abca2 | 2645 | 0.280 | 0.1077 | No |
| 16 | Idh2 | 2666 | 0.279 | 0.1176 | No |
| 17 | Aqp9 | 2753 | 0.270 | 0.1218 | No |
| 18 | Dio2 | 2765 | 0.269 | 0.1321 | No |
| 19 | Pfkm | 2942 | 0.252 | 0.1282 | No |
| 20 | Phyh | 3035 | 0.243 | 0.1308 | No |
| 21 | Nedd4 | 3165 | 0.231 | 0.1299 | No |
| 22 | Cyp27a1 | 3262 | 0.224 | 0.1314 | No |
| 23 | Abca5 | 3447 | 0.209 | 0.1251 | No |
| 24 | Slc22a18 | 3587 | 0.196 | 0.1219 | No |
| 25 | Pex19 | 3917 | 0.171 | 0.1022 | No |
| 26 | Rxra | 4133 | 0.152 | 0.0910 | No |
| 27 | Abcd3 | 4326 | 0.137 | 0.0810 | No |
| 28 | Ar | 4545 | 0.122 | 0.0684 | No |
| 29 | Cat | 4809 | 0.103 | 0.0512 | No |
| 30 | Pex16 | 4810 | 0.102 | 0.0555 | No |
| 31 | Cyp39a1 | 5055 | 0.086 | 0.0392 | No |
| 32 | Slc29a1 | 5366 | 0.065 | 0.0166 | No |
| 33 | Lipe | 5401 | 0.061 | 0.0164 | No |
| 34 | Lonp2 | 5493 | 0.056 | 0.0113 | No |
| 35 | Acsl1 | 5495 | 0.056 | 0.0136 | No |
| 36 | Fads2 | 5717 | 0.041 | -0.0027 | No |
| 37 | Soat2 | 6550 | -0.016 | -0.0698 | No |
| 38 | Fdxr | 7069 | -0.051 | -0.1098 | No |
| 39 | Abcd1 | 7301 | -0.067 | -0.1258 | No |
| 40 | Pex1 | 7860 | -0.106 | -0.1668 | No |
| 41 | Pnpla8 | 8083 | -0.121 | -0.1799 | No |
| 42 | Optn | 8407 | -0.143 | -0.2002 | No |
| 43 | Pex12 | 8537 | -0.151 | -0.2045 | No |
| 44 | Abca4 | 8541 | -0.151 | -0.1985 | No |
| 45 | Hsd17b11 | 8809 | -0.171 | -0.2131 | No |
| 46 | Dhcr24 | 9047 | -0.189 | -0.2246 | No |
| 47 | Nr3c2 | 9226 | -0.201 | -0.2307 | No |
| 48 | Aldh9a1 | 9307 | -0.208 | -0.2286 | No |
| 49 | Pex13 | 9365 | -0.213 | -0.2244 | No |
| 50 | Prdx5 | 9770 | -0.246 | -0.2471 | No |
| 51 | Pxmp2 | 9781 | -0.246 | -0.2377 | No |
| 52 | Pex26 | 9859 | -0.252 | -0.2335 | No |
| 53 | Cyp7b1 | 9939 | -0.259 | -0.2292 | No |
| 54 | Fads1 | 9949 | -0.259 | -0.2192 | No |
| 55 | Atxn1 | 10061 | -0.267 | -0.2171 | No |
| 56 | Pecr | 10219 | -0.281 | -0.2182 | No |
| 57 | Amacr | 10783 | -0.342 | -0.2499 | No |
| 58 | Pex6 | 11169 | -0.393 | -0.2649 | Yes |
| 59 | Abcd2 | 11179 | -0.393 | -0.2493 | Yes |
| 60 | Slc23a2 | 11237 | -0.402 | -0.2373 | Yes |
| 61 | Hsd3b7 | 11256 | -0.404 | -0.2220 | Yes |
| 62 | Slc35b2 | 11341 | -0.420 | -0.2114 | Yes |
| 63 | Npc1 | 11497 | -0.446 | -0.2056 | Yes |
| 64 | Scp2 | 11506 | -0.448 | -0.1876 | Yes |
| 65 | Aldh1a1 | 11516 | -0.450 | -0.1697 | Yes |
| 66 | Retsat | 11677 | -0.480 | -0.1628 | Yes |
| 67 | Gnpat | 11886 | -0.541 | -0.1573 | Yes |
| 68 | Abca1 | 11958 | -0.565 | -0.1396 | Yes |
| 69 | Pex11g | 12059 | -0.606 | -0.1226 | Yes |
| 70 | Idh1 | 12130 | -0.650 | -0.1014 | Yes |
| 71 | Nudt12 | 12154 | -0.674 | -0.0753 | Yes |
| 72 | Crot | 12174 | -0.690 | -0.0482 | Yes |
| 73 | Gclm | 12222 | -0.732 | -0.0217 | Yes |
| 74 | Hsd17b4 | 12272 | -0.798 | 0.0074 | Yes |