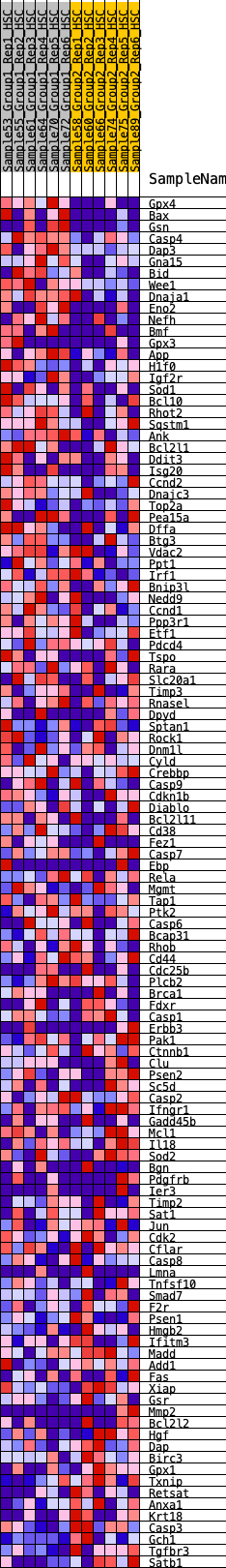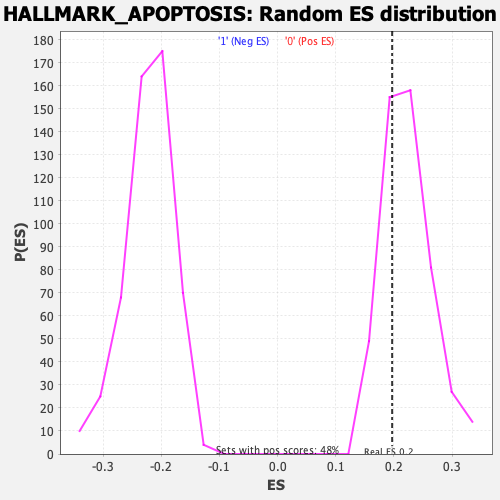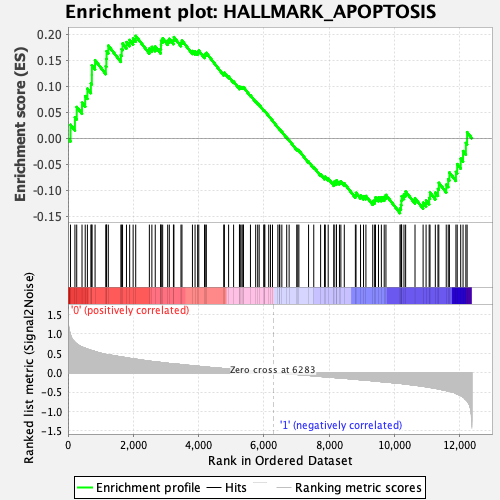
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group2.HSC_Pheno.cls#Group1_versus_Group2_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_APOPTOSIS |
| Enrichment Score (ES) | 0.19686466 |
| Normalized Enrichment Score (NES) | 0.88678974 |
| Nominal p-value | 0.6714876 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gpx4 | 76 | 0.987 | 0.0259 | Yes |
| 2 | Bax | 212 | 0.785 | 0.0404 | Yes |
| 3 | Gsn | 267 | 0.742 | 0.0602 | Yes |
| 4 | Casp4 | 429 | 0.660 | 0.0686 | Yes |
| 5 | Dap3 | 527 | 0.629 | 0.0811 | Yes |
| 6 | Gna15 | 594 | 0.606 | 0.0955 | Yes |
| 7 | Bid | 701 | 0.576 | 0.1056 | Yes |
| 8 | Wee1 | 730 | 0.570 | 0.1218 | Yes |
| 9 | Dnaja1 | 731 | 0.570 | 0.1404 | Yes |
| 10 | Eno2 | 829 | 0.542 | 0.1501 | Yes |
| 11 | Nefh | 1158 | 0.471 | 0.1386 | Yes |
| 12 | Bmf | 1174 | 0.469 | 0.1527 | Yes |
| 13 | Gpx3 | 1181 | 0.467 | 0.1674 | Yes |
| 14 | App | 1234 | 0.461 | 0.1782 | Yes |
| 15 | H1f0 | 1619 | 0.407 | 0.1600 | Yes |
| 16 | Igf2r | 1643 | 0.404 | 0.1713 | Yes |
| 17 | Sod1 | 1670 | 0.399 | 0.1822 | Yes |
| 18 | Bcl10 | 1790 | 0.382 | 0.1849 | Yes |
| 19 | Rhot2 | 1888 | 0.369 | 0.1890 | Yes |
| 20 | Sqstm1 | 1994 | 0.355 | 0.1920 | Yes |
| 21 | Ank | 2073 | 0.346 | 0.1969 | Yes |
| 22 | Bcl2l1 | 2491 | 0.297 | 0.1725 | No |
| 23 | Ddit3 | 2568 | 0.289 | 0.1757 | No |
| 24 | Isg20 | 2671 | 0.279 | 0.1764 | No |
| 25 | Ccnd2 | 2836 | 0.263 | 0.1716 | No |
| 26 | Dnajc3 | 2846 | 0.262 | 0.1794 | No |
| 27 | Top2a | 2850 | 0.261 | 0.1876 | No |
| 28 | Pea15a | 2898 | 0.256 | 0.1921 | No |
| 29 | Dffa | 3052 | 0.241 | 0.1875 | No |
| 30 | Btg3 | 3102 | 0.236 | 0.1912 | No |
| 31 | Vdac2 | 3228 | 0.226 | 0.1883 | No |
| 32 | Ppt1 | 3243 | 0.225 | 0.1945 | No |
| 33 | Irf1 | 3459 | 0.209 | 0.1837 | No |
| 34 | Bnip3l | 3489 | 0.205 | 0.1881 | No |
| 35 | Nedd9 | 3810 | 0.179 | 0.1677 | No |
| 36 | Ccnd1 | 3890 | 0.173 | 0.1669 | No |
| 37 | Ppp3r1 | 3969 | 0.166 | 0.1660 | No |
| 38 | Etf1 | 4004 | 0.163 | 0.1685 | No |
| 39 | Pdcd4 | 4180 | 0.148 | 0.1590 | No |
| 40 | Tspo | 4200 | 0.147 | 0.1622 | No |
| 41 | Rara | 4235 | 0.144 | 0.1642 | No |
| 42 | Slc20a1 | 4768 | 0.105 | 0.1242 | No |
| 43 | Timp3 | 4788 | 0.104 | 0.1260 | No |
| 44 | Rnasel | 4917 | 0.097 | 0.1187 | No |
| 45 | Dpyd | 5070 | 0.085 | 0.1090 | No |
| 46 | Sptan1 | 5249 | 0.073 | 0.0969 | No |
| 47 | Rock1 | 5251 | 0.073 | 0.0991 | No |
| 48 | Dnm1l | 5288 | 0.070 | 0.0985 | No |
| 49 | Cyld | 5327 | 0.068 | 0.0976 | No |
| 50 | Crebbp | 5358 | 0.066 | 0.0973 | No |
| 51 | Casp9 | 5372 | 0.064 | 0.0983 | No |
| 52 | Cdkn1b | 5586 | 0.050 | 0.0826 | No |
| 53 | Diablo | 5746 | 0.039 | 0.0708 | No |
| 54 | Bcl2l11 | 5808 | 0.035 | 0.0670 | No |
| 55 | Cd38 | 5853 | 0.031 | 0.0644 | No |
| 56 | Fez1 | 5993 | 0.020 | 0.0537 | No |
| 57 | Casp7 | 6009 | 0.019 | 0.0531 | No |
| 58 | Ebp | 6027 | 0.018 | 0.0523 | No |
| 59 | Rela | 6145 | 0.010 | 0.0431 | No |
| 60 | Mgmt | 6199 | 0.006 | 0.0389 | No |
| 61 | Tap1 | 6261 | 0.002 | 0.0340 | No |
| 62 | Ptk2 | 6425 | -0.008 | 0.0210 | No |
| 63 | Casp6 | 6471 | -0.011 | 0.0176 | No |
| 64 | Bcap31 | 6485 | -0.012 | 0.0170 | No |
| 65 | Rhob | 6547 | -0.016 | 0.0125 | No |
| 66 | Cd44 | 6696 | -0.025 | 0.0012 | No |
| 67 | Cdc25b | 6770 | -0.030 | -0.0038 | No |
| 68 | Plcb2 | 7003 | -0.046 | -0.0212 | No |
| 69 | Brca1 | 7036 | -0.048 | -0.0222 | No |
| 70 | Fdxr | 7069 | -0.051 | -0.0232 | No |
| 71 | Casp1 | 7365 | -0.071 | -0.0450 | No |
| 72 | Erbb3 | 7524 | -0.081 | -0.0552 | No |
| 73 | Pak1 | 7734 | -0.098 | -0.0691 | No |
| 74 | Ctnnb1 | 7854 | -0.105 | -0.0754 | No |
| 75 | Clu | 7877 | -0.108 | -0.0737 | No |
| 76 | Psen2 | 7966 | -0.112 | -0.0772 | No |
| 77 | Sc5d | 8141 | -0.125 | -0.0874 | No |
| 78 | Casp2 | 8144 | -0.125 | -0.0835 | No |
| 79 | Ifngr1 | 8216 | -0.129 | -0.0851 | No |
| 80 | Gadd45b | 8218 | -0.129 | -0.0809 | No |
| 81 | Mcl1 | 8311 | -0.136 | -0.0840 | No |
| 82 | Il18 | 8354 | -0.140 | -0.0829 | No |
| 83 | Sod2 | 8459 | -0.146 | -0.0867 | No |
| 84 | Bgn | 8798 | -0.170 | -0.1087 | No |
| 85 | Pdgfrb | 8818 | -0.172 | -0.1047 | No |
| 86 | Ier3 | 8956 | -0.182 | -0.1100 | No |
| 87 | Timp2 | 9048 | -0.189 | -0.1113 | No |
| 88 | Sat1 | 9121 | -0.194 | -0.1108 | No |
| 89 | Jun | 9330 | -0.210 | -0.1210 | No |
| 90 | Cdk2 | 9388 | -0.215 | -0.1186 | No |
| 91 | Cflar | 9414 | -0.217 | -0.1136 | No |
| 92 | Casp8 | 9506 | -0.225 | -0.1137 | No |
| 93 | Lmna | 9594 | -0.232 | -0.1133 | No |
| 94 | Tnfsf10 | 9681 | -0.240 | -0.1125 | No |
| 95 | Smad7 | 9734 | -0.243 | -0.1088 | No |
| 96 | F2r | 10164 | -0.277 | -0.1348 | No |
| 97 | Psen1 | 10194 | -0.280 | -0.1281 | No |
| 98 | Hmgb2 | 10207 | -0.280 | -0.1200 | No |
| 99 | Ifitm3 | 10215 | -0.281 | -0.1114 | No |
| 100 | Madd | 10286 | -0.288 | -0.1077 | No |
| 101 | Add1 | 10337 | -0.294 | -0.1023 | No |
| 102 | Fas | 10624 | -0.323 | -0.1151 | No |
| 103 | Xiap | 10872 | -0.354 | -0.1237 | No |
| 104 | Gsr | 10963 | -0.363 | -0.1193 | No |
| 105 | Mmp2 | 11057 | -0.378 | -0.1146 | No |
| 106 | Bcl2l2 | 11083 | -0.381 | -0.1042 | No |
| 107 | Hgf | 11246 | -0.403 | -0.1043 | No |
| 108 | Dap | 11327 | -0.416 | -0.0973 | No |
| 109 | Birc3 | 11353 | -0.422 | -0.0856 | No |
| 110 | Gpx1 | 11580 | -0.462 | -0.0890 | No |
| 111 | Txnip | 11641 | -0.473 | -0.0785 | No |
| 112 | Retsat | 11677 | -0.480 | -0.0657 | No |
| 113 | Anxa1 | 11877 | -0.538 | -0.0644 | No |
| 114 | Krt18 | 11916 | -0.551 | -0.0496 | No |
| 115 | Casp3 | 12023 | -0.589 | -0.0391 | No |
| 116 | Gch1 | 12095 | -0.626 | -0.0245 | No |
| 117 | Tgfbr3 | 12181 | -0.696 | -0.0088 | No |
| 118 | Satb1 | 12219 | -0.725 | 0.0118 | No |