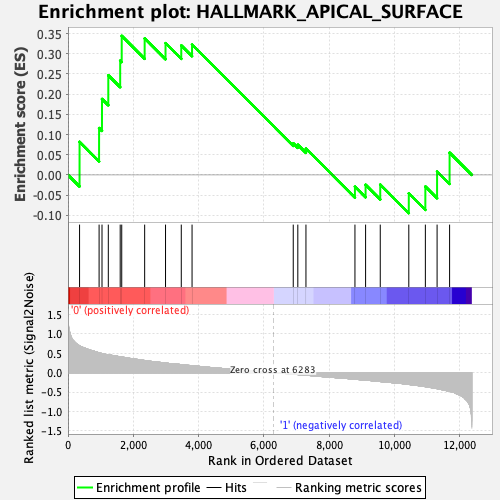
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | HSC.HSC_Pheno.cls#Group1_versus_Group2.HSC_Pheno.cls#Group1_versus_Group2_repos |
| Phenotype | HSC_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_APICAL_SURFACE |
| Enrichment Score (ES) | 0.344204 |
| Normalized Enrichment Score (NES) | 1.1241348 |
| Nominal p-value | 0.28629857 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.974 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Shroom2 | 354 | 0.688 | 0.0819 | Yes |
| 2 | B4galt1 | 952 | 0.513 | 0.1161 | Yes |
| 3 | Cx3cl1 | 1041 | 0.492 | 0.1880 | Yes |
| 4 | App | 1234 | 0.461 | 0.2466 | Yes |
| 5 | Tmem8b | 1598 | 0.410 | 0.2831 | Yes |
| 6 | Ephb4 | 1645 | 0.404 | 0.3442 | Yes |
| 7 | Il2rg | 2347 | 0.314 | 0.3379 | No |
| 8 | Ncoa6 | 2986 | 0.248 | 0.3261 | No |
| 9 | Crocc | 3468 | 0.208 | 0.3205 | No |
| 10 | Dcbld2 | 3798 | 0.179 | 0.3227 | No |
| 11 | Akap7 | 6896 | -0.040 | 0.0782 | No |
| 12 | Brca1 | 7036 | -0.048 | 0.0748 | No |
| 13 | Flot2 | 7285 | -0.066 | 0.0653 | No |
| 14 | Lyn | 8785 | -0.169 | -0.0290 | No |
| 15 | Crybg1 | 9111 | -0.193 | -0.0243 | No |
| 16 | Plaur | 9560 | -0.229 | -0.0238 | No |
| 17 | Mdga1 | 10434 | -0.304 | -0.0456 | No |
| 18 | Adam10 | 10941 | -0.361 | -0.0286 | No |
| 19 | Gata3 | 11300 | -0.412 | 0.0086 | No |
| 20 | Adipor2 | 11683 | -0.482 | 0.0551 | No |

