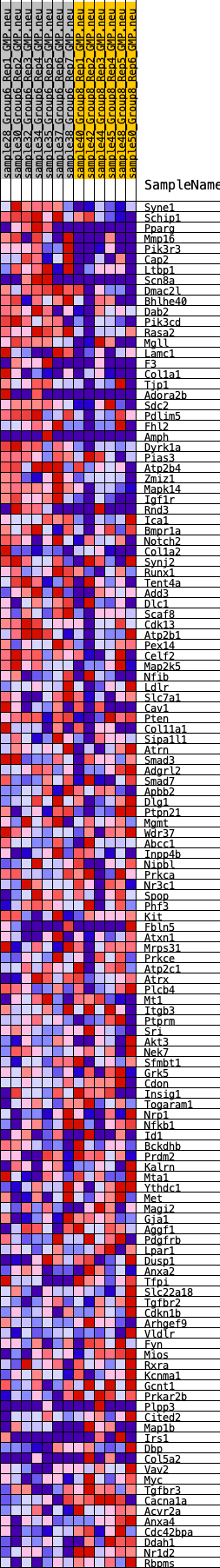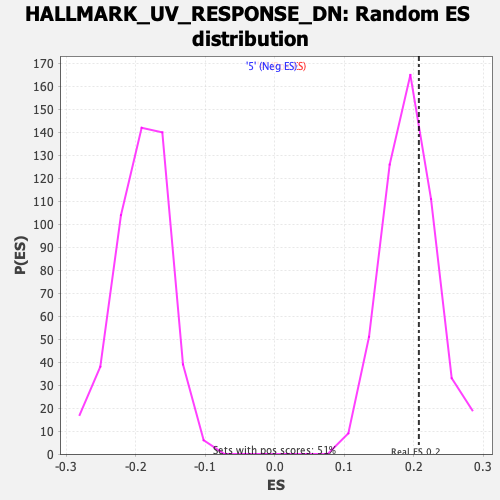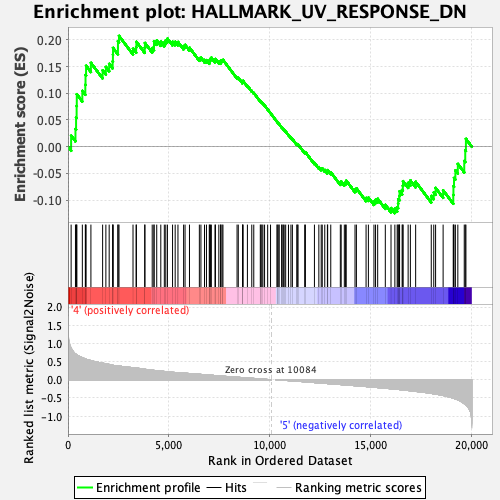
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group6_versus_Group8.GMP.neu_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | GMP.neu_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_UV_RESPONSE_DN |
| Enrichment Score (ES) | 0.2075918 |
| Normalized Enrichment Score (NES) | 1.070593 |
| Nominal p-value | 0.32684824 |
| FDR q-value | 0.90577245 |
| FWER p-Value | 0.987 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Syne1 | 155 | 0.864 | 0.0208 | Yes |
| 2 | Schip1 | 370 | 0.707 | 0.0334 | Yes |
| 3 | Pparg | 401 | 0.699 | 0.0549 | Yes |
| 4 | Mmp16 | 428 | 0.683 | 0.0762 | Yes |
| 5 | Pik3r3 | 436 | 0.680 | 0.0983 | Yes |
| 6 | Cap2 | 711 | 0.607 | 0.1046 | Yes |
| 7 | Ltbp1 | 859 | 0.573 | 0.1161 | Yes |
| 8 | Scn8a | 869 | 0.570 | 0.1345 | Yes |
| 9 | Dmac2l | 897 | 0.563 | 0.1517 | Yes |
| 10 | Bhlhe40 | 1136 | 0.524 | 0.1571 | Yes |
| 11 | Dab2 | 1717 | 0.458 | 0.1431 | Yes |
| 12 | Pik3cd | 1878 | 0.438 | 0.1495 | Yes |
| 13 | Rasa2 | 2042 | 0.421 | 0.1552 | Yes |
| 14 | Mgll | 2214 | 0.398 | 0.1598 | Yes |
| 15 | Lamc1 | 2228 | 0.397 | 0.1722 | Yes |
| 16 | F3 | 2233 | 0.396 | 0.1851 | Yes |
| 17 | Col1a1 | 2477 | 0.377 | 0.1853 | Yes |
| 18 | Tjp1 | 2478 | 0.377 | 0.1978 | Yes |
| 19 | Adora2b | 2529 | 0.373 | 0.2076 | Yes |
| 20 | Sdc2 | 3224 | 0.329 | 0.1836 | No |
| 21 | Pdlim5 | 3379 | 0.321 | 0.1865 | No |
| 22 | Fhl2 | 3391 | 0.320 | 0.1965 | No |
| 23 | Amph | 3805 | 0.289 | 0.1853 | No |
| 24 | Dyrk1a | 3815 | 0.288 | 0.1943 | No |
| 25 | Pias3 | 4176 | 0.263 | 0.1849 | No |
| 26 | Atp2b4 | 4265 | 0.257 | 0.1890 | No |
| 27 | Zmiz1 | 4266 | 0.257 | 0.1975 | No |
| 28 | Mapk14 | 4400 | 0.249 | 0.1990 | No |
| 29 | Igf1r | 4602 | 0.237 | 0.1967 | No |
| 30 | Rnd3 | 4777 | 0.228 | 0.1955 | No |
| 31 | Ica1 | 4849 | 0.224 | 0.1993 | No |
| 32 | Bmpr1a | 4931 | 0.219 | 0.2025 | No |
| 33 | Notch2 | 5185 | 0.204 | 0.1965 | No |
| 34 | Col1a2 | 5315 | 0.200 | 0.1966 | No |
| 35 | Synj2 | 5453 | 0.196 | 0.1962 | No |
| 36 | Runx1 | 5733 | 0.186 | 0.1883 | No |
| 37 | Tent4a | 5805 | 0.182 | 0.1908 | No |
| 38 | Add3 | 6024 | 0.171 | 0.1854 | No |
| 39 | Dlc1 | 6514 | 0.151 | 0.1659 | No |
| 40 | Scaf8 | 6592 | 0.146 | 0.1668 | No |
| 41 | Cdk13 | 6769 | 0.138 | 0.1625 | No |
| 42 | Atp2b1 | 6866 | 0.135 | 0.1622 | No |
| 43 | Pex14 | 7006 | 0.129 | 0.1594 | No |
| 44 | Celf2 | 7029 | 0.128 | 0.1626 | No |
| 45 | Map2k5 | 7064 | 0.126 | 0.1650 | No |
| 46 | Nfib | 7109 | 0.124 | 0.1669 | No |
| 47 | Ldlr | 7290 | 0.115 | 0.1617 | No |
| 48 | Slc7a1 | 7313 | 0.114 | 0.1643 | No |
| 49 | Cav1 | 7480 | 0.106 | 0.1595 | No |
| 50 | Pten | 7566 | 0.103 | 0.1586 | No |
| 51 | Col11a1 | 7580 | 0.102 | 0.1613 | No |
| 52 | Sipa1l1 | 7640 | 0.099 | 0.1617 | No |
| 53 | Atrn | 7684 | 0.098 | 0.1627 | No |
| 54 | Smad3 | 8384 | 0.069 | 0.1299 | No |
| 55 | Adgrl2 | 8445 | 0.067 | 0.1291 | No |
| 56 | Smad7 | 8654 | 0.059 | 0.1206 | No |
| 57 | Apbb2 | 8674 | 0.058 | 0.1215 | No |
| 58 | Dlg1 | 8675 | 0.058 | 0.1234 | No |
| 59 | Ptpn21 | 8897 | 0.049 | 0.1139 | No |
| 60 | Mgmt | 9114 | 0.040 | 0.1044 | No |
| 61 | Wdr37 | 9213 | 0.036 | 0.1007 | No |
| 62 | Abcc1 | 9537 | 0.023 | 0.0852 | No |
| 63 | Inpp4b | 9569 | 0.021 | 0.0844 | No |
| 64 | Nipbl | 9615 | 0.020 | 0.0828 | No |
| 65 | Prkca | 9665 | 0.018 | 0.0809 | No |
| 66 | Nr3c1 | 9748 | 0.014 | 0.0772 | No |
| 67 | Spop | 9902 | 0.007 | 0.0698 | No |
| 68 | Phf3 | 10044 | 0.002 | 0.0627 | No |
| 69 | Kit | 10369 | -0.003 | 0.0465 | No |
| 70 | Fbln5 | 10375 | -0.003 | 0.0464 | No |
| 71 | Atxn1 | 10458 | -0.006 | 0.0425 | No |
| 72 | Mrps31 | 10466 | -0.006 | 0.0423 | No |
| 73 | Prkce | 10595 | -0.011 | 0.0362 | No |
| 74 | Atp2c1 | 10619 | -0.011 | 0.0355 | No |
| 75 | Atrx | 10679 | -0.013 | 0.0329 | No |
| 76 | Plcb4 | 10720 | -0.015 | 0.0314 | No |
| 77 | Mt1 | 10795 | -0.017 | 0.0282 | No |
| 78 | Itgb3 | 10934 | -0.022 | 0.0220 | No |
| 79 | Ptprm | 11056 | -0.027 | 0.0169 | No |
| 80 | Sri | 11128 | -0.030 | 0.0143 | No |
| 81 | Akt3 | 11348 | -0.039 | 0.0045 | No |
| 82 | Nek7 | 11370 | -0.039 | 0.0048 | No |
| 83 | Sfmbt1 | 11409 | -0.041 | 0.0043 | No |
| 84 | Grk5 | 11738 | -0.055 | -0.0104 | No |
| 85 | Cdon | 11774 | -0.057 | -0.0103 | No |
| 86 | Insig1 | 12221 | -0.076 | -0.0302 | No |
| 87 | Togaram1 | 12440 | -0.086 | -0.0384 | No |
| 88 | Nrp1 | 12557 | -0.091 | -0.0412 | No |
| 89 | Nfkb1 | 12608 | -0.093 | -0.0406 | No |
| 90 | Id1 | 12733 | -0.099 | -0.0436 | No |
| 91 | Bckdhb | 12864 | -0.104 | -0.0467 | No |
| 92 | Prdm2 | 12878 | -0.104 | -0.0439 | No |
| 93 | Kalrn | 13031 | -0.110 | -0.0479 | No |
| 94 | Mta1 | 13505 | -0.131 | -0.0673 | No |
| 95 | Ythdc1 | 13547 | -0.132 | -0.0650 | No |
| 96 | Met | 13704 | -0.139 | -0.0683 | No |
| 97 | Magi2 | 13765 | -0.141 | -0.0666 | No |
| 98 | Gja1 | 13797 | -0.142 | -0.0635 | No |
| 99 | Aggf1 | 14234 | -0.161 | -0.0801 | No |
| 100 | Pdgfrb | 14303 | -0.164 | -0.0781 | No |
| 101 | Lpar1 | 14782 | -0.186 | -0.0960 | No |
| 102 | Dusp1 | 14897 | -0.192 | -0.0954 | No |
| 103 | Anxa2 | 15173 | -0.205 | -0.1024 | No |
| 104 | Tfpi | 15252 | -0.209 | -0.0994 | No |
| 105 | Slc22a18 | 15358 | -0.214 | -0.0977 | No |
| 106 | Tgfbr2 | 15736 | -0.232 | -0.1089 | No |
| 107 | Cdkn1b | 16019 | -0.247 | -0.1150 | No |
| 108 | Arhgef9 | 16210 | -0.258 | -0.1160 | No |
| 109 | Vldlr | 16325 | -0.262 | -0.1131 | No |
| 110 | Fyn | 16371 | -0.265 | -0.1066 | No |
| 111 | Mios | 16377 | -0.265 | -0.0981 | No |
| 112 | Rxra | 16436 | -0.268 | -0.0921 | No |
| 113 | Kcnma1 | 16443 | -0.268 | -0.0836 | No |
| 114 | Gcnt1 | 16570 | -0.274 | -0.0808 | No |
| 115 | Prkar2b | 16605 | -0.277 | -0.0734 | No |
| 116 | Plpp3 | 16614 | -0.278 | -0.0646 | No |
| 117 | Cited2 | 16868 | -0.294 | -0.0676 | No |
| 118 | Map1b | 16979 | -0.301 | -0.0632 | No |
| 119 | Irs1 | 17241 | -0.319 | -0.0657 | No |
| 120 | Dbp | 18015 | -0.374 | -0.0922 | No |
| 121 | Col5a2 | 18139 | -0.385 | -0.0857 | No |
| 122 | Vav2 | 18230 | -0.393 | -0.0772 | No |
| 123 | Myc | 18603 | -0.431 | -0.0817 | No |
| 124 | Tgfbr3 | 19107 | -0.505 | -0.0903 | No |
| 125 | Cacna1a | 19115 | -0.506 | -0.0739 | No |
| 126 | Acvr2a | 19144 | -0.513 | -0.0584 | No |
| 127 | Anxa4 | 19211 | -0.525 | -0.0443 | No |
| 128 | Cdc42bpa | 19333 | -0.550 | -0.0323 | No |
| 129 | Ddah1 | 19651 | -0.656 | -0.0265 | No |
| 130 | Nr1d2 | 19701 | -0.677 | -0.0066 | No |
| 131 | Rbpms | 19734 | -0.694 | 0.0147 | No |