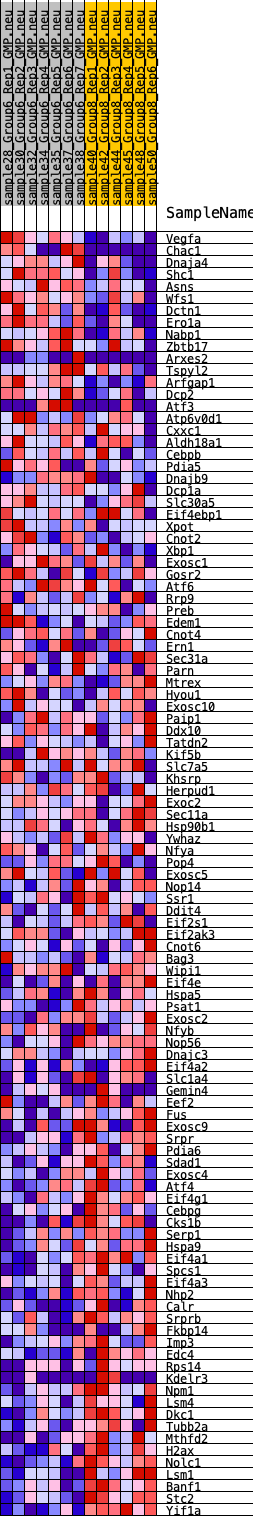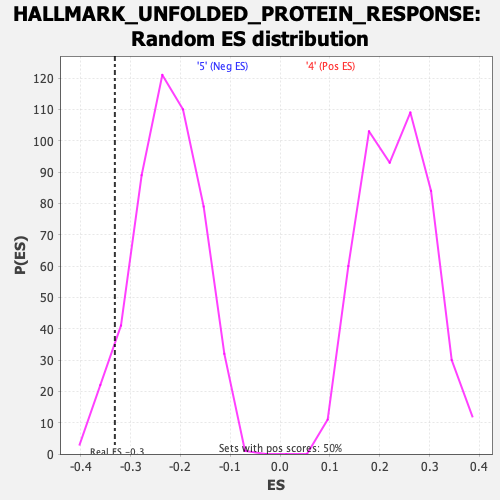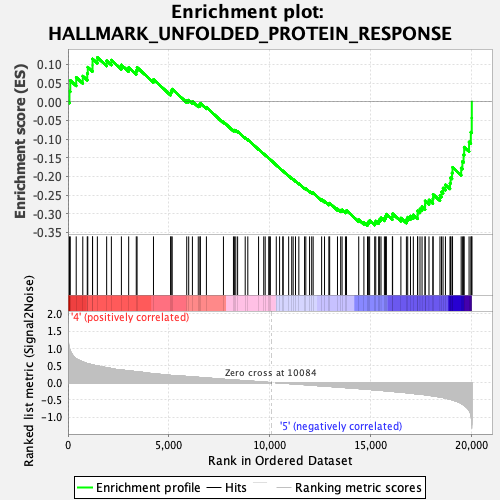
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group6_versus_Group8.GMP.neu_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | GMP.neu_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | -0.3316269 |
| Normalized Enrichment Score (NES) | -1.4707958 |
| Nominal p-value | 0.05421687 |
| FDR q-value | 0.13849236 |
| FWER p-Value | 0.424 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Vegfa | 73 | 0.978 | 0.0285 | No |
| 2 | Chac1 | 113 | 0.925 | 0.0570 | No |
| 3 | Dnaja4 | 407 | 0.695 | 0.0651 | No |
| 4 | Shc1 | 733 | 0.602 | 0.0686 | No |
| 5 | Asns | 955 | 0.552 | 0.0757 | No |
| 6 | Wfs1 | 981 | 0.550 | 0.0925 | No |
| 7 | Dctn1 | 1217 | 0.515 | 0.0977 | No |
| 8 | Ero1a | 1218 | 0.515 | 0.1146 | No |
| 9 | Nabp1 | 1456 | 0.483 | 0.1186 | No |
| 10 | Zbtb17 | 1917 | 0.434 | 0.1098 | No |
| 11 | Arxes2 | 2151 | 0.407 | 0.1114 | No |
| 12 | Tspyl2 | 2648 | 0.367 | 0.0986 | No |
| 13 | Arfgap1 | 3009 | 0.341 | 0.0918 | No |
| 14 | Dcp2 | 3385 | 0.321 | 0.0835 | No |
| 15 | Atf3 | 3425 | 0.317 | 0.0920 | No |
| 16 | Atp6v0d1 | 4239 | 0.259 | 0.0597 | No |
| 17 | Cxxc1 | 5088 | 0.209 | 0.0240 | No |
| 18 | Aldh18a1 | 5117 | 0.208 | 0.0295 | No |
| 19 | Cebpb | 5167 | 0.205 | 0.0338 | No |
| 20 | Pdia5 | 5888 | 0.177 | 0.0034 | No |
| 21 | Dnajb9 | 5986 | 0.172 | 0.0042 | No |
| 22 | Dcp1a | 6170 | 0.169 | 0.0006 | No |
| 23 | Slc30a5 | 6460 | 0.153 | -0.0089 | No |
| 24 | Eif4ebp1 | 6540 | 0.149 | -0.0079 | No |
| 25 | Xpot | 6564 | 0.148 | -0.0042 | No |
| 26 | Cnot2 | 6864 | 0.135 | -0.0148 | No |
| 27 | Xbp1 | 7709 | 0.097 | -0.0540 | No |
| 28 | Exosc1 | 8205 | 0.077 | -0.0763 | No |
| 29 | Gosr2 | 8251 | 0.075 | -0.0761 | No |
| 30 | Atf6 | 8321 | 0.072 | -0.0772 | No |
| 31 | Rrp9 | 8412 | 0.068 | -0.0795 | No |
| 32 | Preb | 8788 | 0.053 | -0.0965 | No |
| 33 | Edem1 | 8917 | 0.048 | -0.1014 | No |
| 34 | Cnot4 | 9451 | 0.026 | -0.1273 | No |
| 35 | Ern1 | 9707 | 0.015 | -0.1396 | No |
| 36 | Sec31a | 9779 | 0.013 | -0.1427 | No |
| 37 | Parn | 9958 | 0.005 | -0.1515 | No |
| 38 | Mtrex | 9984 | 0.004 | -0.1526 | No |
| 39 | Hyou1 | 10036 | 0.002 | -0.1551 | No |
| 40 | Exosc10 | 10329 | -0.001 | -0.1697 | No |
| 41 | Paip1 | 10493 | -0.007 | -0.1777 | No |
| 42 | Ddx10 | 10651 | -0.012 | -0.1852 | No |
| 43 | Tatdn2 | 10671 | -0.013 | -0.1857 | No |
| 44 | Kif5b | 10943 | -0.023 | -0.1986 | No |
| 45 | Slc7a5 | 11095 | -0.029 | -0.2052 | No |
| 46 | Khsrp | 11166 | -0.032 | -0.2077 | No |
| 47 | Herpud1 | 11283 | -0.037 | -0.2123 | No |
| 48 | Exoc2 | 11448 | -0.043 | -0.2191 | No |
| 49 | Sec11a | 11731 | -0.054 | -0.2314 | No |
| 50 | Hsp90b1 | 11794 | -0.057 | -0.2327 | No |
| 51 | Ywhaz | 11980 | -0.065 | -0.2398 | No |
| 52 | Nfya | 12086 | -0.070 | -0.2428 | No |
| 53 | Pop4 | 12158 | -0.073 | -0.2439 | No |
| 54 | Exosc5 | 12578 | -0.092 | -0.2620 | No |
| 55 | Nop14 | 12722 | -0.099 | -0.2659 | No |
| 56 | Ssr1 | 12937 | -0.107 | -0.2731 | No |
| 57 | Ddit4 | 12977 | -0.108 | -0.2715 | No |
| 58 | Eif2s1 | 13364 | -0.124 | -0.2868 | No |
| 59 | Eif2ak3 | 13523 | -0.131 | -0.2905 | No |
| 60 | Cnot6 | 13592 | -0.134 | -0.2895 | No |
| 61 | Bag3 | 13760 | -0.141 | -0.2932 | No |
| 62 | Wipi1 | 13812 | -0.143 | -0.2910 | No |
| 63 | Eif4e | 14418 | -0.169 | -0.3158 | No |
| 64 | Hspa5 | 14674 | -0.181 | -0.3227 | No |
| 65 | Psat1 | 14853 | -0.190 | -0.3254 | Yes |
| 66 | Exosc2 | 14896 | -0.192 | -0.3212 | Yes |
| 67 | Nfyb | 14959 | -0.195 | -0.3179 | Yes |
| 68 | Nop56 | 15210 | -0.206 | -0.3236 | Yes |
| 69 | Dnajc3 | 15262 | -0.209 | -0.3193 | Yes |
| 70 | Eif4a2 | 15406 | -0.216 | -0.3194 | Yes |
| 71 | Slc1a4 | 15455 | -0.219 | -0.3146 | Yes |
| 72 | Gemin4 | 15523 | -0.222 | -0.3107 | Yes |
| 73 | Eef2 | 15691 | -0.230 | -0.3115 | Yes |
| 74 | Fus | 15745 | -0.233 | -0.3065 | Yes |
| 75 | Exosc9 | 15790 | -0.235 | -0.3009 | Yes |
| 76 | Srpr | 16087 | -0.251 | -0.3075 | Yes |
| 77 | Pdia6 | 16099 | -0.252 | -0.2998 | Yes |
| 78 | Sdad1 | 16509 | -0.273 | -0.3113 | Yes |
| 79 | Exosc4 | 16782 | -0.289 | -0.3155 | Yes |
| 80 | Atf4 | 16847 | -0.294 | -0.3090 | Yes |
| 81 | Eif4g1 | 16985 | -0.302 | -0.3060 | Yes |
| 82 | Cebpg | 17126 | -0.312 | -0.3027 | Yes |
| 83 | Cks1b | 17339 | -0.325 | -0.3027 | Yes |
| 84 | Serp1 | 17340 | -0.325 | -0.2920 | Yes |
| 85 | Hspa9 | 17456 | -0.332 | -0.2869 | Yes |
| 86 | Eif4a1 | 17558 | -0.338 | -0.2808 | Yes |
| 87 | Spcs1 | 17708 | -0.349 | -0.2768 | Yes |
| 88 | Eif4a3 | 17710 | -0.349 | -0.2654 | Yes |
| 89 | Nhp2 | 17902 | -0.365 | -0.2630 | Yes |
| 90 | Calr | 18096 | -0.381 | -0.2601 | Yes |
| 91 | Srprb | 18106 | -0.383 | -0.2480 | Yes |
| 92 | Fkbp14 | 18447 | -0.413 | -0.2515 | Yes |
| 93 | Imp3 | 18525 | -0.422 | -0.2415 | Yes |
| 94 | Edc4 | 18600 | -0.431 | -0.2310 | Yes |
| 95 | Rps14 | 18718 | -0.445 | -0.2222 | Yes |
| 96 | Kdelr3 | 18944 | -0.477 | -0.2178 | Yes |
| 97 | Npm1 | 18973 | -0.481 | -0.2034 | Yes |
| 98 | Lsm4 | 19044 | -0.493 | -0.1907 | Yes |
| 99 | Dkc1 | 19070 | -0.499 | -0.1755 | Yes |
| 100 | Tubb2a | 19498 | -0.598 | -0.1773 | Yes |
| 101 | Mthfd2 | 19559 | -0.618 | -0.1600 | Yes |
| 102 | H2ax | 19618 | -0.645 | -0.1417 | Yes |
| 103 | Nolc1 | 19647 | -0.653 | -0.1216 | Yes |
| 104 | Lsm1 | 19890 | -0.794 | -0.1076 | Yes |
| 105 | Banf1 | 19978 | -0.927 | -0.0815 | Yes |
| 106 | Stc2 | 20023 | -1.219 | -0.0436 | Yes |
| 107 | Yif1a | 20026 | -1.328 | 0.0000 | Yes |