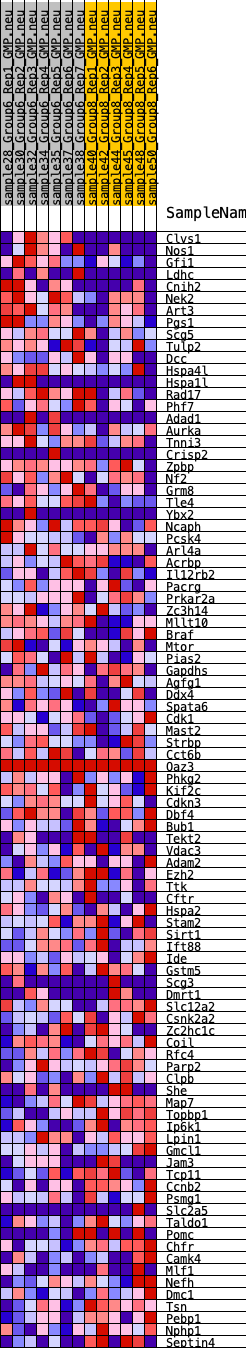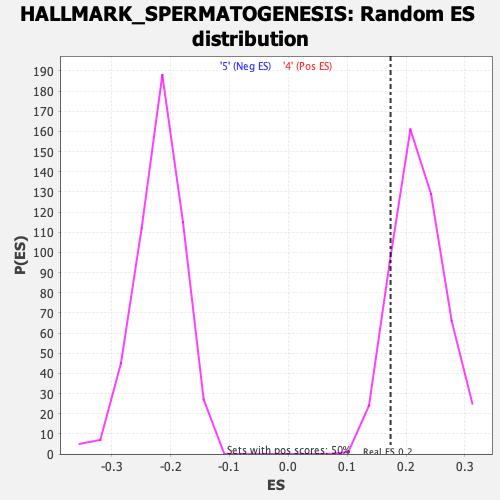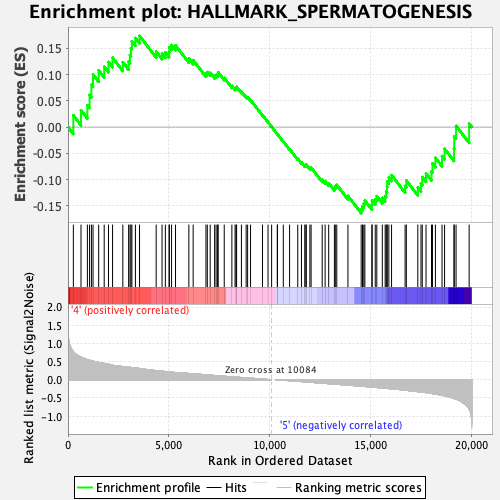
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group6_versus_Group8.GMP.neu_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | GMP.neu_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.17367198 |
| Normalized Enrichment Score (NES) | 0.78853273 |
| Nominal p-value | 0.86427146 |
| FDR q-value | 0.86030763 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Clvs1 | 265 | 0.773 | 0.0224 | Yes |
| 2 | Nos1 | 645 | 0.623 | 0.0321 | Yes |
| 3 | Gfi1 | 961 | 0.552 | 0.0417 | Yes |
| 4 | Ldhc | 1069 | 0.536 | 0.0611 | Yes |
| 5 | Cnih2 | 1161 | 0.520 | 0.0805 | Yes |
| 6 | Nek2 | 1241 | 0.511 | 0.1001 | Yes |
| 7 | Art3 | 1521 | 0.476 | 0.1081 | Yes |
| 8 | Pgs1 | 1795 | 0.447 | 0.1150 | Yes |
| 9 | Scg5 | 2009 | 0.424 | 0.1239 | Yes |
| 10 | Tulp2 | 2210 | 0.398 | 0.1323 | Yes |
| 11 | Dcc | 2719 | 0.362 | 0.1235 | Yes |
| 12 | Hspa4l | 3006 | 0.342 | 0.1249 | Yes |
| 13 | Hspa1l | 3074 | 0.339 | 0.1372 | Yes |
| 14 | Rad17 | 3123 | 0.337 | 0.1503 | Yes |
| 15 | Phf7 | 3168 | 0.333 | 0.1635 | Yes |
| 16 | Adad1 | 3343 | 0.322 | 0.1696 | Yes |
| 17 | Aurka | 3547 | 0.309 | 0.1737 | Yes |
| 18 | Tnni3 | 4373 | 0.250 | 0.1438 | No |
| 19 | Crisp2 | 4664 | 0.234 | 0.1401 | No |
| 20 | Zpbp | 4828 | 0.225 | 0.1423 | No |
| 21 | Nf2 | 5013 | 0.214 | 0.1429 | No |
| 22 | Grm8 | 5025 | 0.213 | 0.1522 | No |
| 23 | Tle4 | 5136 | 0.206 | 0.1562 | No |
| 24 | Ybx2 | 5332 | 0.199 | 0.1556 | No |
| 25 | Ncaph | 5993 | 0.172 | 0.1304 | No |
| 26 | Pcsk4 | 6207 | 0.167 | 0.1274 | No |
| 27 | Arl4a | 6841 | 0.135 | 0.1019 | No |
| 28 | Acrbp | 6909 | 0.133 | 0.1047 | No |
| 29 | Il12rb2 | 7051 | 0.127 | 0.1035 | No |
| 30 | Pacrg | 7272 | 0.116 | 0.0978 | No |
| 31 | Prkar2a | 7345 | 0.112 | 0.0994 | No |
| 32 | Zc3h14 | 7419 | 0.109 | 0.1007 | No |
| 33 | Mllt10 | 7448 | 0.108 | 0.1043 | No |
| 34 | Braf | 7748 | 0.095 | 0.0937 | No |
| 35 | Mtor | 8127 | 0.080 | 0.0784 | No |
| 36 | Pias2 | 8285 | 0.073 | 0.0739 | No |
| 37 | Gapdhs | 8336 | 0.071 | 0.0746 | No |
| 38 | Agfg1 | 8361 | 0.070 | 0.0767 | No |
| 39 | Ddx4 | 8600 | 0.060 | 0.0675 | No |
| 40 | Spata6 | 8835 | 0.052 | 0.0582 | No |
| 41 | Cdk1 | 8908 | 0.049 | 0.0568 | No |
| 42 | Mast2 | 9053 | 0.043 | 0.0516 | No |
| 43 | Strbp | 9646 | 0.019 | 0.0227 | No |
| 44 | Cct6b | 9920 | 0.006 | 0.0093 | No |
| 45 | Oaz3 | 10098 | 0.000 | 0.0004 | No |
| 46 | Phkg2 | 10383 | -0.003 | -0.0137 | No |
| 47 | Kif2c | 10385 | -0.003 | -0.0136 | No |
| 48 | Cdkn3 | 10677 | -0.013 | -0.0276 | No |
| 49 | Dbf4 | 10989 | -0.025 | -0.0420 | No |
| 50 | Bub1 | 11394 | -0.041 | -0.0604 | No |
| 51 | Tekt2 | 11578 | -0.048 | -0.0674 | No |
| 52 | Vdac3 | 11742 | -0.055 | -0.0730 | No |
| 53 | Adam2 | 11800 | -0.058 | -0.0732 | No |
| 54 | Ezh2 | 11815 | -0.058 | -0.0713 | No |
| 55 | Ttk | 11995 | -0.066 | -0.0772 | No |
| 56 | Cftr | 12056 | -0.069 | -0.0770 | No |
| 57 | Hspa2 | 12607 | -0.093 | -0.1003 | No |
| 58 | Stam2 | 12751 | -0.100 | -0.1029 | No |
| 59 | Sirt1 | 12928 | -0.106 | -0.1068 | No |
| 60 | Ift88 | 13211 | -0.118 | -0.1155 | No |
| 61 | Ide | 13256 | -0.120 | -0.1122 | No |
| 62 | Gstm5 | 13329 | -0.122 | -0.1102 | No |
| 63 | Scg3 | 13883 | -0.147 | -0.1312 | No |
| 64 | Dmrt1 | 14539 | -0.175 | -0.1560 | No |
| 65 | Slc12a2 | 14605 | -0.178 | -0.1510 | No |
| 66 | Csnk2a2 | 14669 | -0.181 | -0.1458 | No |
| 67 | Zc2hc1c | 14722 | -0.184 | -0.1400 | No |
| 68 | Coil | 15073 | -0.201 | -0.1483 | No |
| 69 | Rfc4 | 15082 | -0.201 | -0.1394 | No |
| 70 | Parp2 | 15237 | -0.208 | -0.1375 | No |
| 71 | Clpb | 15314 | -0.212 | -0.1316 | No |
| 72 | She | 15587 | -0.225 | -0.1348 | No |
| 73 | Map7 | 15729 | -0.232 | -0.1312 | No |
| 74 | Topbp1 | 15779 | -0.234 | -0.1229 | No |
| 75 | Ip6k1 | 15820 | -0.237 | -0.1139 | No |
| 76 | Lpin1 | 15829 | -0.237 | -0.1034 | No |
| 77 | Gmcl1 | 15913 | -0.241 | -0.0964 | No |
| 78 | Jam3 | 16046 | -0.249 | -0.0916 | No |
| 79 | Tcp11 | 16717 | -0.284 | -0.1120 | No |
| 80 | Ccnb2 | 16779 | -0.289 | -0.1018 | No |
| 81 | Psmg1 | 17348 | -0.325 | -0.1153 | No |
| 82 | Slc2a5 | 17505 | -0.335 | -0.1076 | No |
| 83 | Taldo1 | 17569 | -0.338 | -0.0952 | No |
| 84 | Pomc | 17755 | -0.352 | -0.0882 | No |
| 85 | Chfr | 18028 | -0.375 | -0.0845 | No |
| 86 | Camk4 | 18075 | -0.380 | -0.0693 | No |
| 87 | Mlf1 | 18221 | -0.392 | -0.0585 | No |
| 88 | Nefh | 18549 | -0.424 | -0.0553 | No |
| 89 | Dmc1 | 18675 | -0.439 | -0.0413 | No |
| 90 | Tsn | 19142 | -0.513 | -0.0411 | No |
| 91 | Pebp1 | 19154 | -0.515 | -0.0179 | No |
| 92 | Nphp1 | 19245 | -0.533 | 0.0022 | No |
| 93 | Septin4 | 19894 | -0.800 | 0.0066 | No |