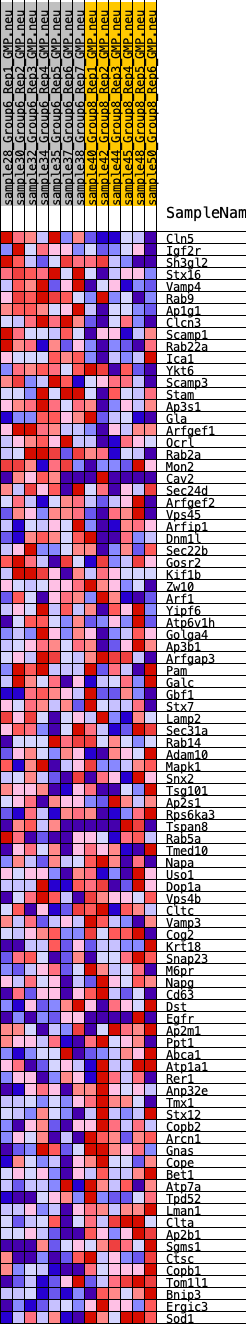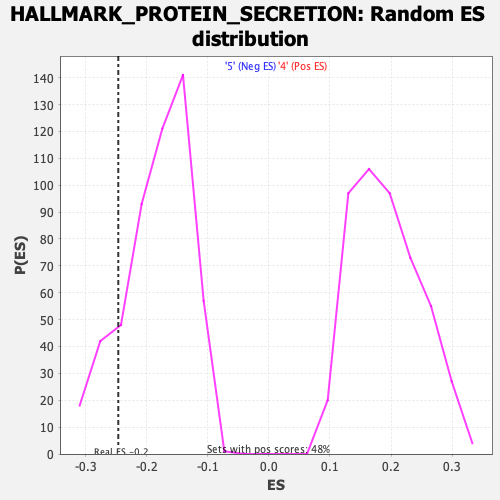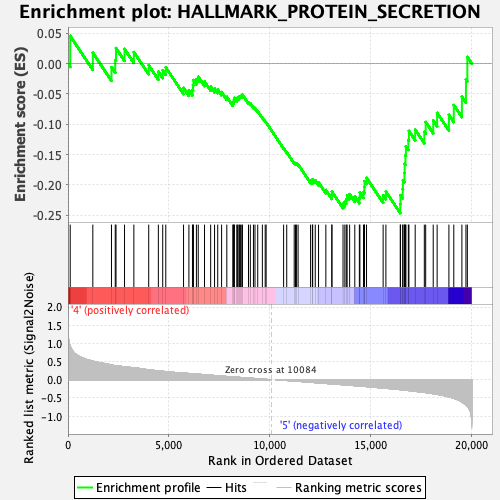
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group6_versus_Group8.GMP.neu_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | GMP.neu_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | -0.24649377 |
| Normalized Enrichment Score (NES) | -1.3431697 |
| Nominal p-value | 0.15163147 |
| FDR q-value | 0.2046669 |
| FWER p-Value | 0.665 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cln5 | 110 | 0.926 | 0.0458 | No |
| 2 | Igf2r | 1231 | 0.513 | 0.0181 | No |
| 3 | Sh3gl2 | 2156 | 0.406 | -0.0058 | No |
| 4 | Stx16 | 2343 | 0.386 | 0.0063 | No |
| 5 | Vamp4 | 2381 | 0.383 | 0.0257 | No |
| 6 | Rab9 | 2803 | 0.356 | 0.0243 | No |
| 7 | Ap1g1 | 3266 | 0.325 | 0.0192 | No |
| 8 | Clcn3 | 4003 | 0.272 | -0.0026 | No |
| 9 | Scamp1 | 4481 | 0.243 | -0.0131 | No |
| 10 | Rab22a | 4697 | 0.233 | -0.0109 | No |
| 11 | Ica1 | 4849 | 0.224 | -0.0061 | No |
| 12 | Ykt6 | 5730 | 0.186 | -0.0399 | No |
| 13 | Scamp3 | 5996 | 0.172 | -0.0437 | No |
| 14 | Stam | 6172 | 0.168 | -0.0431 | No |
| 15 | Ap3s1 | 6200 | 0.167 | -0.0352 | No |
| 16 | Gla | 6220 | 0.166 | -0.0270 | No |
| 17 | Arfgef1 | 6368 | 0.158 | -0.0256 | No |
| 18 | Ocrl | 6459 | 0.153 | -0.0216 | No |
| 19 | Rab2a | 6770 | 0.138 | -0.0295 | No |
| 20 | Mon2 | 7075 | 0.126 | -0.0377 | No |
| 21 | Cav2 | 7268 | 0.116 | -0.0409 | No |
| 22 | Sec24d | 7425 | 0.109 | -0.0427 | No |
| 23 | Arfgef2 | 7617 | 0.100 | -0.0467 | No |
| 24 | Vps45 | 7876 | 0.089 | -0.0547 | No |
| 25 | Arfip1 | 8173 | 0.078 | -0.0652 | No |
| 26 | Dnm1l | 8204 | 0.077 | -0.0625 | No |
| 27 | Sec22b | 8234 | 0.076 | -0.0597 | No |
| 28 | Gosr2 | 8251 | 0.075 | -0.0564 | No |
| 29 | Kif1b | 8373 | 0.070 | -0.0586 | No |
| 30 | Zw10 | 8400 | 0.068 | -0.0561 | No |
| 31 | Arf1 | 8474 | 0.066 | -0.0561 | No |
| 32 | Yipf6 | 8500 | 0.065 | -0.0537 | No |
| 33 | Atp6v1h | 8559 | 0.062 | -0.0532 | No |
| 34 | Golga4 | 8620 | 0.060 | -0.0529 | No |
| 35 | Ap3b1 | 8651 | 0.059 | -0.0511 | No |
| 36 | Arfgap3 | 8951 | 0.047 | -0.0635 | No |
| 37 | Pam | 9042 | 0.043 | -0.0656 | No |
| 38 | Galc | 9197 | 0.037 | -0.0713 | No |
| 39 | Gbf1 | 9273 | 0.034 | -0.0732 | No |
| 40 | Stx7 | 9409 | 0.028 | -0.0784 | No |
| 41 | Lamp2 | 9639 | 0.019 | -0.0889 | No |
| 42 | Sec31a | 9779 | 0.013 | -0.0951 | No |
| 43 | Rab14 | 9826 | 0.011 | -0.0968 | No |
| 44 | Adam10 | 10693 | -0.014 | -0.1395 | No |
| 45 | Mapk1 | 10847 | -0.019 | -0.1461 | No |
| 46 | Snx2 | 11218 | -0.033 | -0.1629 | No |
| 47 | Tsg101 | 11282 | -0.037 | -0.1640 | No |
| 48 | Ap2s1 | 11336 | -0.038 | -0.1645 | No |
| 49 | Rps6ka3 | 11411 | -0.041 | -0.1660 | No |
| 50 | Tspan8 | 12030 | -0.068 | -0.1932 | No |
| 51 | Rab5a | 12120 | -0.072 | -0.1937 | No |
| 52 | Tmed10 | 12135 | -0.072 | -0.1904 | No |
| 53 | Napa | 12262 | -0.078 | -0.1924 | No |
| 54 | Uso1 | 12425 | -0.085 | -0.1958 | No |
| 55 | Dop1a | 12787 | -0.101 | -0.2083 | No |
| 56 | Vps4b | 13086 | -0.112 | -0.2170 | No |
| 57 | Cltc | 13087 | -0.112 | -0.2108 | No |
| 58 | Vamp3 | 13639 | -0.137 | -0.2309 | No |
| 59 | Cog2 | 13730 | -0.140 | -0.2276 | No |
| 60 | Krt18 | 13801 | -0.143 | -0.2232 | No |
| 61 | Snap23 | 13842 | -0.144 | -0.2172 | No |
| 62 | M6pr | 13967 | -0.149 | -0.2151 | No |
| 63 | Napg | 14223 | -0.161 | -0.2190 | No |
| 64 | Cd63 | 14449 | -0.171 | -0.2208 | No |
| 65 | Dst | 14476 | -0.172 | -0.2126 | No |
| 66 | Egfr | 14654 | -0.180 | -0.2115 | No |
| 67 | Ap2m1 | 14699 | -0.183 | -0.2036 | No |
| 68 | Ppt1 | 14705 | -0.183 | -0.1937 | No |
| 69 | Abca1 | 14805 | -0.188 | -0.1883 | No |
| 70 | Atp1a1 | 15628 | -0.227 | -0.2169 | No |
| 71 | Rer1 | 15765 | -0.233 | -0.2108 | No |
| 72 | Anp32e | 16478 | -0.271 | -0.2314 | Yes |
| 73 | Tmx1 | 16488 | -0.272 | -0.2168 | Yes |
| 74 | Stx12 | 16599 | -0.277 | -0.2070 | Yes |
| 75 | Copb2 | 16612 | -0.277 | -0.1922 | Yes |
| 76 | Arcn1 | 16686 | -0.281 | -0.1803 | Yes |
| 77 | Gnas | 16696 | -0.282 | -0.1651 | Yes |
| 78 | Cope | 16735 | -0.285 | -0.1511 | Yes |
| 79 | Bet1 | 16758 | -0.287 | -0.1363 | Yes |
| 80 | Atp7a | 16879 | -0.295 | -0.1260 | Yes |
| 81 | Tpd52 | 16907 | -0.297 | -0.1108 | Yes |
| 82 | Lman1 | 17218 | -0.318 | -0.1088 | Yes |
| 83 | Clta | 17671 | -0.345 | -0.1123 | Yes |
| 84 | Ap2b1 | 17738 | -0.351 | -0.0961 | Yes |
| 85 | Sgms1 | 18112 | -0.383 | -0.0936 | Yes |
| 86 | Ctsc | 18306 | -0.400 | -0.0811 | Yes |
| 87 | Copb1 | 18890 | -0.469 | -0.0844 | Yes |
| 88 | Tom1l1 | 19132 | -0.511 | -0.0681 | Yes |
| 89 | Bnip3 | 19534 | -0.609 | -0.0545 | Yes |
| 90 | Ergic3 | 19735 | -0.695 | -0.0260 | Yes |
| 91 | Sod1 | 19805 | -0.730 | 0.0111 | Yes |