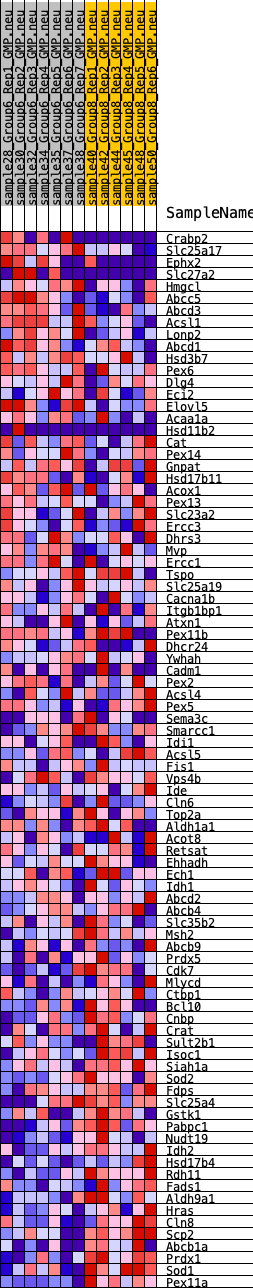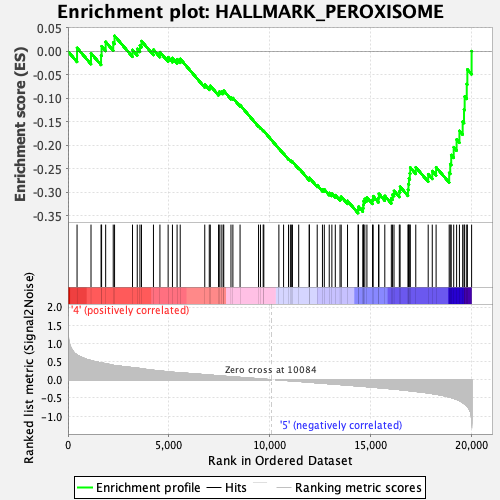
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group6_versus_Group8.GMP.neu_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | GMP.neu_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.34465858 |
| Normalized Enrichment Score (NES) | -1.4691657 |
| Nominal p-value | 0.026104419 |
| FDR q-value | 0.12454588 |
| FWER p-Value | 0.424 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Crabp2 | 449 | 0.676 | 0.0075 | No |
| 2 | Slc25a17 | 1141 | 0.524 | -0.0040 | No |
| 3 | Ephx2 | 1640 | 0.464 | -0.0083 | No |
| 4 | Slc27a2 | 1666 | 0.462 | 0.0109 | No |
| 5 | Hmgcl | 1868 | 0.439 | 0.0203 | No |
| 6 | Abcc5 | 2243 | 0.394 | 0.0190 | No |
| 7 | Abcd3 | 2309 | 0.389 | 0.0330 | No |
| 8 | Acsl1 | 3199 | 0.330 | 0.0031 | No |
| 9 | Lonp2 | 3434 | 0.316 | 0.0054 | No |
| 10 | Abcd1 | 3567 | 0.307 | 0.0123 | No |
| 11 | Hsd3b7 | 3644 | 0.302 | 0.0219 | No |
| 12 | Pex6 | 4238 | 0.259 | 0.0037 | No |
| 13 | Dlg4 | 4559 | 0.240 | -0.0017 | No |
| 14 | Eci2 | 4966 | 0.217 | -0.0124 | No |
| 15 | Elovl5 | 5180 | 0.204 | -0.0141 | No |
| 16 | Acaa1a | 5410 | 0.199 | -0.0167 | No |
| 17 | Hsd11b2 | 5565 | 0.191 | -0.0160 | No |
| 18 | Cat | 6782 | 0.138 | -0.0709 | No |
| 19 | Pex14 | 7006 | 0.129 | -0.0763 | No |
| 20 | Gnpat | 7057 | 0.126 | -0.0732 | No |
| 21 | Hsd17b11 | 7466 | 0.107 | -0.0890 | No |
| 22 | Acox1 | 7500 | 0.106 | -0.0859 | No |
| 23 | Pex13 | 7594 | 0.101 | -0.0861 | No |
| 24 | Slc23a2 | 7690 | 0.098 | -0.0865 | No |
| 25 | Ercc3 | 7724 | 0.096 | -0.0839 | No |
| 26 | Dhrs3 | 8082 | 0.082 | -0.0982 | No |
| 27 | Mvp | 8177 | 0.078 | -0.0994 | No |
| 28 | Ercc1 | 8533 | 0.063 | -0.1144 | No |
| 29 | Tspo | 9449 | 0.026 | -0.1592 | No |
| 30 | Slc25a19 | 9545 | 0.023 | -0.1629 | No |
| 31 | Cacna1b | 9680 | 0.017 | -0.1689 | No |
| 32 | Itgb1bp1 | 9711 | 0.015 | -0.1697 | No |
| 33 | Atxn1 | 10458 | -0.006 | -0.2069 | No |
| 34 | Pex11b | 10691 | -0.013 | -0.2179 | No |
| 35 | Dhcr24 | 10936 | -0.022 | -0.2292 | No |
| 36 | Ywhah | 11028 | -0.026 | -0.2326 | No |
| 37 | Cadm1 | 11076 | -0.028 | -0.2337 | No |
| 38 | Pex2 | 11131 | -0.030 | -0.2350 | No |
| 39 | Acsl4 | 11441 | -0.043 | -0.2486 | No |
| 40 | Pex5 | 11963 | -0.065 | -0.2719 | No |
| 41 | Sema3c | 11969 | -0.065 | -0.2693 | No |
| 42 | Smarcc1 | 12359 | -0.082 | -0.2851 | No |
| 43 | Idi1 | 12619 | -0.094 | -0.2939 | No |
| 44 | Acsl5 | 12711 | -0.098 | -0.2942 | No |
| 45 | Fis1 | 12955 | -0.107 | -0.3016 | No |
| 46 | Vps4b | 13086 | -0.112 | -0.3032 | No |
| 47 | Ide | 13256 | -0.120 | -0.3063 | No |
| 48 | Cln6 | 13490 | -0.130 | -0.3122 | No |
| 49 | Top2a | 13549 | -0.132 | -0.3093 | No |
| 50 | Aldh1a1 | 13862 | -0.145 | -0.3185 | No |
| 51 | Acot8 | 14385 | -0.168 | -0.3372 | Yes |
| 52 | Retsat | 14408 | -0.169 | -0.3308 | Yes |
| 53 | Ehhadh | 14621 | -0.179 | -0.3336 | Yes |
| 54 | Ech1 | 14649 | -0.180 | -0.3269 | Yes |
| 55 | Idh1 | 14657 | -0.180 | -0.3193 | Yes |
| 56 | Abcd2 | 14720 | -0.184 | -0.3143 | Yes |
| 57 | Abcb4 | 14824 | -0.189 | -0.3111 | Yes |
| 58 | Slc35b2 | 15102 | -0.202 | -0.3160 | Yes |
| 59 | Msh2 | 15134 | -0.203 | -0.3085 | Yes |
| 60 | Abcb9 | 15401 | -0.216 | -0.3123 | Yes |
| 61 | Prdx5 | 15418 | -0.217 | -0.3035 | Yes |
| 62 | Cdk7 | 15711 | -0.231 | -0.3079 | Yes |
| 63 | Mlycd | 16035 | -0.248 | -0.3131 | Yes |
| 64 | Ctbp1 | 16096 | -0.252 | -0.3049 | Yes |
| 65 | Bcl10 | 16168 | -0.255 | -0.2972 | Yes |
| 66 | Cnbp | 16431 | -0.267 | -0.2985 | Yes |
| 67 | Crat | 16469 | -0.271 | -0.2883 | Yes |
| 68 | Sult2b1 | 16860 | -0.294 | -0.2948 | Yes |
| 69 | Isoc1 | 16885 | -0.295 | -0.2829 | Yes |
| 70 | Siah1a | 16909 | -0.297 | -0.2709 | Yes |
| 71 | Sod2 | 16952 | -0.300 | -0.2597 | Yes |
| 72 | Fdps | 16973 | -0.301 | -0.2474 | Yes |
| 73 | Slc25a4 | 17247 | -0.319 | -0.2469 | Yes |
| 74 | Gstk1 | 17863 | -0.361 | -0.2617 | Yes |
| 75 | Pabpc1 | 18065 | -0.379 | -0.2550 | Yes |
| 76 | Nudt19 | 18255 | -0.395 | -0.2469 | Yes |
| 77 | Idh2 | 18904 | -0.470 | -0.2586 | Yes |
| 78 | Hsd17b4 | 18956 | -0.478 | -0.2399 | Yes |
| 79 | Rdh11 | 19005 | -0.486 | -0.2208 | Yes |
| 80 | Fads1 | 19128 | -0.509 | -0.2043 | Yes |
| 81 | Aldh9a1 | 19272 | -0.537 | -0.1877 | Yes |
| 82 | Hras | 19412 | -0.571 | -0.1693 | Yes |
| 83 | Cln8 | 19573 | -0.624 | -0.1496 | Yes |
| 84 | Scp2 | 19640 | -0.652 | -0.1240 | Yes |
| 85 | Abcb1a | 19672 | -0.664 | -0.0961 | Yes |
| 86 | Prdx1 | 19776 | -0.718 | -0.0694 | Yes |
| 87 | Sod1 | 19805 | -0.730 | -0.0385 | Yes |
| 88 | Pex11a | 20016 | -1.116 | 0.0005 | Yes |