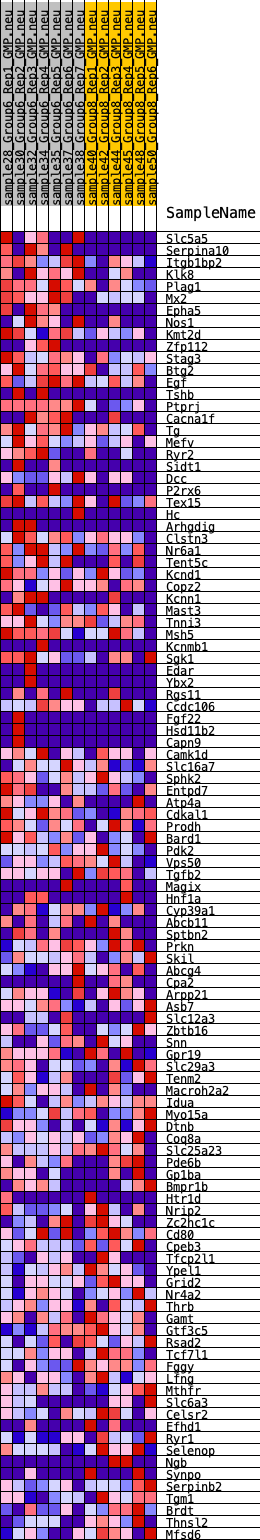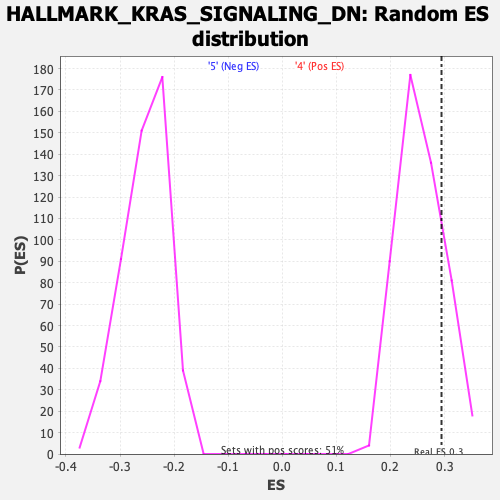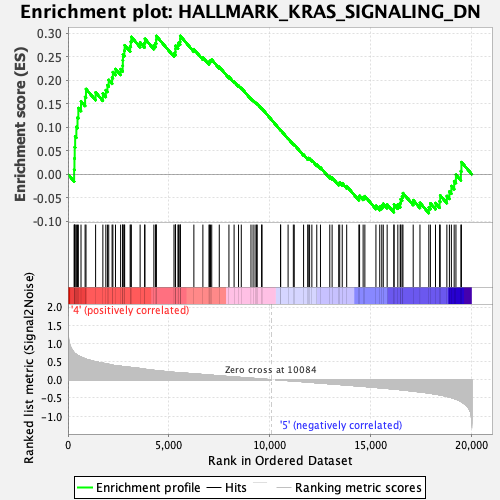
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group6_versus_Group8.GMP.neu_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | GMP.neu_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.29435334 |
| Normalized Enrichment Score (NES) | 1.1452334 |
| Nominal p-value | 0.18972331 |
| FDR q-value | 0.7953933 |
| FWER p-Value | 0.96 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Slc5a5 | 304 | 0.742 | 0.0097 | Yes |
| 2 | Serpina10 | 320 | 0.736 | 0.0338 | Yes |
| 3 | Itgb1bp2 | 333 | 0.723 | 0.0576 | Yes |
| 4 | Klk8 | 355 | 0.715 | 0.0807 | Yes |
| 5 | Plag1 | 415 | 0.691 | 0.1010 | Yes |
| 6 | Mx2 | 472 | 0.669 | 0.1207 | Yes |
| 7 | Epha5 | 513 | 0.658 | 0.1409 | Yes |
| 8 | Nos1 | 645 | 0.623 | 0.1553 | Yes |
| 9 | Kmt2d | 848 | 0.575 | 0.1646 | Yes |
| 10 | Zfp112 | 892 | 0.564 | 0.1814 | Yes |
| 11 | Stag3 | 1369 | 0.494 | 0.1742 | Yes |
| 12 | Btg2 | 1729 | 0.457 | 0.1716 | Yes |
| 13 | Egf | 1876 | 0.438 | 0.1790 | Yes |
| 14 | Tshb | 1956 | 0.430 | 0.1895 | Yes |
| 15 | Ptprj | 2020 | 0.423 | 0.2007 | Yes |
| 16 | Cacna1f | 2187 | 0.401 | 0.2058 | Yes |
| 17 | Tg | 2226 | 0.397 | 0.2173 | Yes |
| 18 | Mefv | 2355 | 0.384 | 0.2239 | Yes |
| 19 | Ryr2 | 2609 | 0.369 | 0.2236 | Yes |
| 20 | Sidt1 | 2712 | 0.363 | 0.2307 | Yes |
| 21 | Dcc | 2719 | 0.362 | 0.2426 | Yes |
| 22 | P2rx6 | 2727 | 0.361 | 0.2544 | Yes |
| 23 | Tex15 | 2794 | 0.357 | 0.2632 | Yes |
| 24 | Hc | 2810 | 0.356 | 0.2744 | Yes |
| 25 | Arhgdig | 3082 | 0.339 | 0.2722 | Yes |
| 26 | Clstn3 | 3116 | 0.338 | 0.2819 | Yes |
| 27 | Nr6a1 | 3139 | 0.336 | 0.2922 | Yes |
| 28 | Tent5c | 3577 | 0.306 | 0.2805 | Yes |
| 29 | Kcnd1 | 3798 | 0.290 | 0.2793 | Yes |
| 30 | Copz2 | 3816 | 0.288 | 0.2881 | Yes |
| 31 | Kcnn1 | 4259 | 0.257 | 0.2746 | Yes |
| 32 | Mast3 | 4343 | 0.253 | 0.2789 | Yes |
| 33 | Tnni3 | 4373 | 0.250 | 0.2859 | Yes |
| 34 | Msh5 | 4376 | 0.250 | 0.2942 | Yes |
| 35 | Kcnmb1 | 5256 | 0.200 | 0.2569 | Yes |
| 36 | Sgk1 | 5326 | 0.199 | 0.2601 | Yes |
| 37 | Edar | 5328 | 0.199 | 0.2668 | Yes |
| 38 | Ybx2 | 5332 | 0.199 | 0.2734 | Yes |
| 39 | Rgs11 | 5456 | 0.196 | 0.2738 | Yes |
| 40 | Ccdc106 | 5475 | 0.195 | 0.2795 | Yes |
| 41 | Fgf22 | 5559 | 0.191 | 0.2817 | Yes |
| 42 | Hsd11b2 | 5565 | 0.191 | 0.2879 | Yes |
| 43 | Capn9 | 5566 | 0.191 | 0.2944 | Yes |
| 44 | Camk1d | 6242 | 0.165 | 0.2660 | No |
| 45 | Slc16a7 | 6689 | 0.142 | 0.2484 | No |
| 46 | Sphk2 | 6992 | 0.129 | 0.2376 | No |
| 47 | Entpd7 | 7022 | 0.128 | 0.2405 | No |
| 48 | Atp4a | 7068 | 0.126 | 0.2425 | No |
| 49 | Cdkal1 | 7128 | 0.123 | 0.2437 | No |
| 50 | Prodh | 7502 | 0.106 | 0.2285 | No |
| 51 | Bard1 | 7981 | 0.086 | 0.2074 | No |
| 52 | Pdk2 | 8237 | 0.076 | 0.1971 | No |
| 53 | Vps50 | 8457 | 0.066 | 0.1884 | No |
| 54 | Tgfb2 | 8594 | 0.061 | 0.1836 | No |
| 55 | Magix | 9074 | 0.042 | 0.1610 | No |
| 56 | Hnf1a | 9178 | 0.038 | 0.1571 | No |
| 57 | Cyp39a1 | 9277 | 0.034 | 0.1533 | No |
| 58 | Abcb11 | 9352 | 0.030 | 0.1506 | No |
| 59 | Sptbn2 | 9383 | 0.029 | 0.1501 | No |
| 60 | Prkn | 9599 | 0.020 | 0.1400 | No |
| 61 | Skil | 9623 | 0.019 | 0.1395 | No |
| 62 | Abcg4 | 10539 | -0.009 | 0.0938 | No |
| 63 | Cpa2 | 10547 | -0.009 | 0.0938 | No |
| 64 | Arpp21 | 10915 | -0.021 | 0.0760 | No |
| 65 | Asb7 | 11175 | -0.032 | 0.0641 | No |
| 66 | Slc12a3 | 11220 | -0.033 | 0.0630 | No |
| 67 | Zbtb16 | 11683 | -0.053 | 0.0416 | No |
| 68 | Snn | 11893 | -0.061 | 0.0332 | No |
| 69 | Gpr19 | 11908 | -0.062 | 0.0346 | No |
| 70 | Slc29a3 | 11981 | -0.065 | 0.0332 | No |
| 71 | Tenm2 | 12094 | -0.071 | 0.0299 | No |
| 72 | Macroh2a2 | 12337 | -0.081 | 0.0205 | No |
| 73 | Idua | 12521 | -0.089 | 0.0143 | No |
| 74 | Myo15a | 12982 | -0.108 | -0.0051 | No |
| 75 | Dtnb | 13098 | -0.112 | -0.0071 | No |
| 76 | Coq8a | 13425 | -0.127 | -0.0192 | No |
| 77 | Slc25a23 | 13475 | -0.129 | -0.0173 | No |
| 78 | Pde6b | 13600 | -0.134 | -0.0190 | No |
| 79 | Gp1ba | 13821 | -0.143 | -0.0252 | No |
| 80 | Bmpr1b | 14426 | -0.170 | -0.0498 | No |
| 81 | Htr1d | 14460 | -0.171 | -0.0457 | No |
| 82 | Nrip2 | 14636 | -0.179 | -0.0485 | No |
| 83 | Zc2hc1c | 14722 | -0.184 | -0.0465 | No |
| 84 | Cd80 | 15271 | -0.209 | -0.0670 | No |
| 85 | Cpeb3 | 15460 | -0.219 | -0.0690 | No |
| 86 | Tfcp2l1 | 15561 | -0.223 | -0.0665 | No |
| 87 | Ypel1 | 15635 | -0.227 | -0.0625 | No |
| 88 | Grid2 | 15828 | -0.237 | -0.0642 | No |
| 89 | Nr4a2 | 16162 | -0.255 | -0.0723 | No |
| 90 | Thrb | 16169 | -0.255 | -0.0640 | No |
| 91 | Gamt | 16357 | -0.264 | -0.0644 | No |
| 92 | Gtf3c5 | 16474 | -0.271 | -0.0611 | No |
| 93 | Rsad2 | 16497 | -0.273 | -0.0530 | No |
| 94 | Tcf7l1 | 16567 | -0.274 | -0.0473 | No |
| 95 | Fggy | 16611 | -0.277 | -0.0401 | No |
| 96 | Lfng | 17119 | -0.311 | -0.0550 | No |
| 97 | Mthfr | 17455 | -0.332 | -0.0607 | No |
| 98 | Slc6a3 | 17894 | -0.364 | -0.0704 | No |
| 99 | Celsr2 | 17972 | -0.371 | -0.0617 | No |
| 100 | Efhd1 | 18225 | -0.392 | -0.0611 | No |
| 101 | Ryr1 | 18428 | -0.411 | -0.0574 | No |
| 102 | Selenop | 18458 | -0.414 | -0.0449 | No |
| 103 | Ngb | 18789 | -0.455 | -0.0461 | No |
| 104 | Synpo | 18915 | -0.472 | -0.0365 | No |
| 105 | Serpinb2 | 19013 | -0.486 | -0.0250 | No |
| 106 | Tgm1 | 19151 | -0.514 | -0.0145 | No |
| 107 | Brdt | 19239 | -0.531 | -0.0010 | No |
| 108 | Thnsl2 | 19485 | -0.595 | 0.0068 | No |
| 109 | Mfsd6 | 19511 | -0.603 | 0.0259 | No |