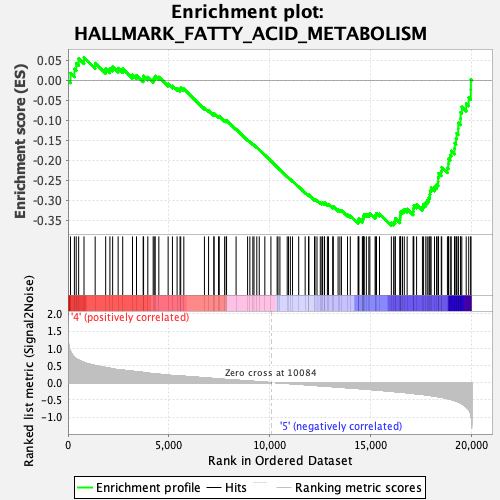
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group6_versus_Group8.GMP.neu_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | GMP.neu_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
| Enrichment Score (ES) | -0.36369112 |
| Normalized Enrichment Score (NES) | -1.5242536 |
| Nominal p-value | 0.018292682 |
| FDR q-value | 0.1110371 |
| FWER p-Value | 0.332 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nthl1 | 124 | 0.901 | 0.0183 | No |
| 2 | D2hgdh | 317 | 0.736 | 0.0287 | No |
| 3 | Ube2l6 | 408 | 0.694 | 0.0431 | No |
| 4 | Cd1d2 | 528 | 0.654 | 0.0550 | No |
| 5 | Acads | 794 | 0.587 | 0.0577 | No |
| 6 | Aoc3 | 1347 | 0.498 | 0.0435 | No |
| 7 | Hmgcl | 1868 | 0.439 | 0.0293 | No |
| 8 | Hmgcs2 | 2079 | 0.417 | 0.0302 | No |
| 9 | Mgll | 2214 | 0.398 | 0.0343 | No |
| 10 | Hsdl2 | 2486 | 0.376 | 0.0309 | No |
| 11 | Acot2 | 2713 | 0.363 | 0.0294 | No |
| 12 | Acsl1 | 3199 | 0.330 | 0.0140 | No |
| 13 | Idh3b | 3397 | 0.320 | 0.0128 | No |
| 14 | Nsdhl | 3728 | 0.296 | 0.0043 | No |
| 15 | Decr1 | 3748 | 0.295 | 0.0114 | No |
| 16 | Metap1 | 3958 | 0.275 | 0.0084 | No |
| 17 | Pcbd1 | 4224 | 0.261 | 0.0022 | No |
| 18 | Gpd2 | 4275 | 0.256 | 0.0066 | No |
| 19 | Rdh1 | 4325 | 0.254 | 0.0111 | No |
| 20 | Pts | 4505 | 0.241 | 0.0087 | No |
| 21 | Eci2 | 4966 | 0.217 | -0.0086 | No |
| 22 | Elovl5 | 5180 | 0.204 | -0.0137 | No |
| 23 | Acaa1a | 5410 | 0.199 | -0.0198 | No |
| 24 | Eno2 | 5556 | 0.191 | -0.0219 | No |
| 25 | Sms-ps | 5590 | 0.191 | -0.0184 | No |
| 26 | Acadvl | 5744 | 0.185 | -0.0210 | No |
| 27 | Bphl | 6766 | 0.139 | -0.0686 | No |
| 28 | Rap1gds1 | 6971 | 0.130 | -0.0753 | No |
| 29 | Slc22a5 | 7234 | 0.118 | -0.0852 | No |
| 30 | Ncaph2 | 7243 | 0.117 | -0.0824 | No |
| 31 | Hsd17b11 | 7466 | 0.107 | -0.0907 | No |
| 32 | Acox1 | 7500 | 0.106 | -0.0895 | No |
| 33 | Acsm3 | 7767 | 0.094 | -0.1003 | No |
| 34 | Cpt2 | 7845 | 0.090 | -0.1017 | No |
| 35 | Aco2 | 7863 | 0.090 | -0.1001 | No |
| 36 | Gapdhs | 8336 | 0.071 | -0.1219 | No |
| 37 | Sdha | 8906 | 0.049 | -0.1492 | No |
| 38 | Hibch | 9019 | 0.044 | -0.1536 | No |
| 39 | Hccs | 9166 | 0.038 | -0.1599 | No |
| 40 | Dlst | 9224 | 0.036 | -0.1618 | No |
| 41 | Hmgcs1 | 9363 | 0.030 | -0.1679 | No |
| 42 | Xist | 9485 | 0.025 | -0.1733 | No |
| 43 | Acaa2 | 9761 | 0.013 | -0.1867 | No |
| 44 | Prdx6 | 10063 | 0.001 | -0.2018 | No |
| 45 | Aldh3a2 | 10371 | -0.003 | -0.2172 | No |
| 46 | Adsl | 10415 | -0.005 | -0.2192 | No |
| 47 | Ppara | 10509 | -0.008 | -0.2237 | No |
| 48 | Suclg2 | 10871 | -0.020 | -0.2413 | No |
| 49 | Dhcr24 | 10936 | -0.022 | -0.2439 | No |
| 50 | Ywhah | 11028 | -0.026 | -0.2478 | No |
| 51 | Odc1 | 11135 | -0.031 | -0.2523 | No |
| 52 | Acsl4 | 11441 | -0.043 | -0.2664 | No |
| 53 | Sdhc | 11761 | -0.056 | -0.2809 | No |
| 54 | Grhpr | 11929 | -0.063 | -0.2876 | No |
| 55 | Auh | 11947 | -0.064 | -0.2867 | No |
| 56 | Gpd1 | 12225 | -0.076 | -0.2986 | No |
| 57 | Fasn | 12250 | -0.077 | -0.2977 | No |
| 58 | Pdhb | 12348 | -0.081 | -0.3003 | No |
| 59 | Cbr1 | 12512 | -0.089 | -0.3061 | No |
| 60 | Kmt5a | 12590 | -0.092 | -0.3075 | No |
| 61 | Idi1 | 12619 | -0.094 | -0.3063 | No |
| 62 | Acsl5 | 12711 | -0.098 | -0.3082 | No |
| 63 | Acadl | 12723 | -0.099 | -0.3061 | No |
| 64 | Bckdhb | 12864 | -0.104 | -0.3103 | No |
| 65 | Trp53inp2 | 12917 | -0.106 | -0.3100 | No |
| 66 | Aldoa | 13127 | -0.114 | -0.3174 | No |
| 67 | Hadhb | 13157 | -0.116 | -0.3157 | No |
| 68 | Glul | 13394 | -0.125 | -0.3242 | No |
| 69 | Gad2 | 13484 | -0.130 | -0.3251 | No |
| 70 | Sucla2 | 13561 | -0.132 | -0.3253 | No |
| 71 | Aldh1a1 | 13862 | -0.145 | -0.3364 | No |
| 72 | H2az1 | 14000 | -0.151 | -0.3392 | No |
| 73 | Acot8 | 14385 | -0.168 | -0.3539 | No |
| 74 | Retsat | 14408 | -0.169 | -0.3504 | No |
| 75 | Bmpr1b | 14426 | -0.170 | -0.3467 | No |
| 76 | Eno3 | 14598 | -0.177 | -0.3504 | No |
| 77 | Ehhadh | 14621 | -0.179 | -0.3467 | No |
| 78 | Ech1 | 14649 | -0.180 | -0.3431 | No |
| 79 | Idh1 | 14657 | -0.180 | -0.3386 | No |
| 80 | Idh3g | 14692 | -0.182 | -0.3353 | No |
| 81 | Cryz | 14796 | -0.187 | -0.3354 | No |
| 82 | Reep6 | 14919 | -0.193 | -0.3363 | No |
| 83 | Acadm | 14971 | -0.196 | -0.3335 | No |
| 84 | Ltc4s | 15230 | -0.207 | -0.3408 | No |
| 85 | Cpt1a | 15264 | -0.209 | -0.3368 | No |
| 86 | Gstz1 | 15300 | -0.211 | -0.3328 | No |
| 87 | Urod | 15448 | -0.218 | -0.3342 | No |
| 88 | Mlycd | 16035 | -0.248 | -0.3569 | Yes |
| 89 | Car2 | 16149 | -0.254 | -0.3557 | Yes |
| 90 | Echs1 | 16219 | -0.259 | -0.3521 | Yes |
| 91 | Hsd17b7 | 16232 | -0.259 | -0.3456 | Yes |
| 92 | Me1 | 16460 | -0.270 | -0.3497 | Yes |
| 93 | Crat | 16469 | -0.271 | -0.3427 | Yes |
| 94 | Hsp90aa1 | 16484 | -0.272 | -0.3360 | Yes |
| 95 | G0s2 | 16499 | -0.273 | -0.3293 | Yes |
| 96 | Nbn | 16588 | -0.276 | -0.3262 | Yes |
| 97 | Dld | 16678 | -0.281 | -0.3230 | Yes |
| 98 | Adipor2 | 16819 | -0.291 | -0.3221 | Yes |
| 99 | Hsd17b10 | 17118 | -0.311 | -0.3286 | Yes |
| 100 | Mcee | 17130 | -0.312 | -0.3207 | Yes |
| 101 | Ephx1 | 17150 | -0.314 | -0.3131 | Yes |
| 102 | Fh1 | 17287 | -0.322 | -0.3111 | Yes |
| 103 | Pdha1 | 17577 | -0.339 | -0.3164 | Yes |
| 104 | Acss1 | 17634 | -0.342 | -0.3099 | Yes |
| 105 | Acat2 | 17744 | -0.351 | -0.3058 | Yes |
| 106 | Mdh2 | 17816 | -0.357 | -0.2997 | Yes |
| 107 | Eci1 | 17898 | -0.364 | -0.2938 | Yes |
| 108 | Gcdh | 17944 | -0.369 | -0.2860 | Yes |
| 109 | Serinc1 | 17957 | -0.370 | -0.2765 | Yes |
| 110 | Hsph1 | 18008 | -0.373 | -0.2688 | Yes |
| 111 | Etfdh | 18176 | -0.388 | -0.2667 | Yes |
| 112 | Uros | 18277 | -0.398 | -0.2608 | Yes |
| 113 | Gabarapl1 | 18354 | -0.404 | -0.2536 | Yes |
| 114 | Apex1 | 18359 | -0.405 | -0.2428 | Yes |
| 115 | Psme1 | 18376 | -0.406 | -0.2325 | Yes |
| 116 | Ugdh | 18511 | -0.420 | -0.2278 | Yes |
| 117 | Maoa | 18534 | -0.423 | -0.2174 | Yes |
| 118 | Ostc | 18816 | -0.459 | -0.2190 | Yes |
| 119 | S100a10 | 18865 | -0.467 | -0.2087 | Yes |
| 120 | Erp29 | 18870 | -0.467 | -0.1962 | Yes |
| 121 | Hsd17b4 | 18956 | -0.478 | -0.1874 | Yes |
| 122 | Rdh11 | 19005 | -0.486 | -0.1766 | Yes |
| 123 | Ldha | 19167 | -0.518 | -0.1706 | Yes |
| 124 | Suclg1 | 19180 | -0.520 | -0.1570 | Yes |
| 125 | Hadh | 19244 | -0.533 | -0.1457 | Yes |
| 126 | Aldh9a1 | 19272 | -0.537 | -0.1324 | Yes |
| 127 | Alad | 19352 | -0.555 | -0.1212 | Yes |
| 128 | Sdhd | 19355 | -0.556 | -0.1062 | Yes |
| 129 | Mif | 19463 | -0.587 | -0.0956 | Yes |
| 130 | Blvra | 19470 | -0.590 | -0.0798 | Yes |
| 131 | Cd36 | 19530 | -0.607 | -0.0662 | Yes |
| 132 | Cbr3 | 19748 | -0.702 | -0.0580 | Yes |
| 133 | Mdh1 | 19872 | -0.784 | -0.0428 | Yes |
| 134 | Lgals1 | 19975 | -0.923 | -0.0227 | Yes |
| 135 | Cpox | 19979 | -0.927 | 0.0024 | Yes |