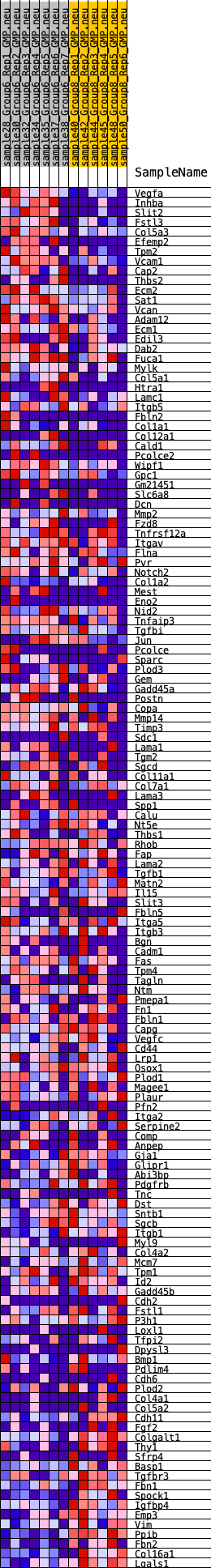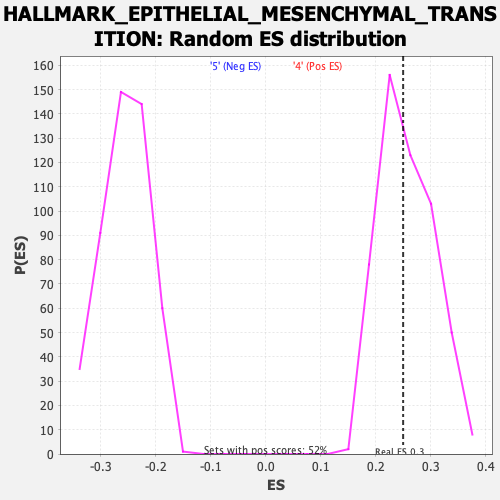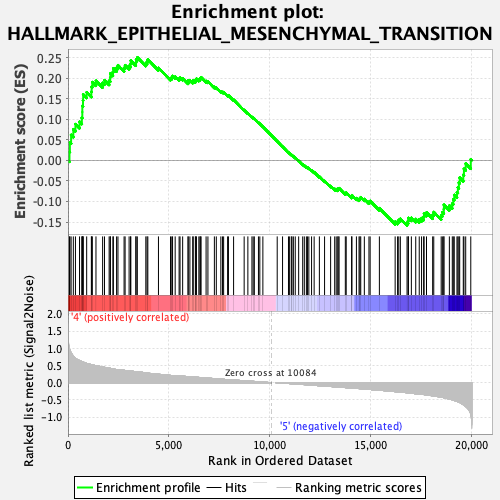
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group6_versus_Group8.GMP.neu_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | GMP.neu_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | 0.2502017 |
| Normalized Enrichment Score (NES) | 0.9761107 |
| Nominal p-value | 0.4923077 |
| FDR q-value | 0.8229402 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Vegfa | 73 | 0.978 | 0.0207 | Yes |
| 2 | Inhba | 93 | 0.946 | 0.0433 | Yes |
| 3 | Slit2 | 154 | 0.865 | 0.0618 | Yes |
| 4 | Fstl3 | 264 | 0.773 | 0.0756 | Yes |
| 5 | Col5a3 | 369 | 0.708 | 0.0880 | Yes |
| 6 | Efemp2 | 572 | 0.640 | 0.0938 | Yes |
| 7 | Tpm2 | 686 | 0.613 | 0.1033 | Yes |
| 8 | Vcam1 | 710 | 0.607 | 0.1173 | Yes |
| 9 | Cap2 | 711 | 0.607 | 0.1324 | Yes |
| 10 | Thbs2 | 744 | 0.599 | 0.1457 | Yes |
| 11 | Ecm2 | 753 | 0.597 | 0.1602 | Yes |
| 12 | Sat1 | 926 | 0.557 | 0.1654 | Yes |
| 13 | Vcan | 1158 | 0.521 | 0.1668 | Yes |
| 14 | Adam12 | 1174 | 0.518 | 0.1789 | Yes |
| 15 | Ecm1 | 1206 | 0.515 | 0.1902 | Yes |
| 16 | Edil3 | 1387 | 0.491 | 0.1933 | Yes |
| 17 | Dab2 | 1717 | 0.458 | 0.1882 | Yes |
| 18 | Fuca1 | 1809 | 0.445 | 0.1947 | Yes |
| 19 | Mylk | 2040 | 0.421 | 0.1936 | Yes |
| 20 | Col5a1 | 2095 | 0.415 | 0.2012 | Yes |
| 21 | Htra1 | 2100 | 0.415 | 0.2114 | Yes |
| 22 | Lamc1 | 2228 | 0.397 | 0.2149 | Yes |
| 23 | Itgb5 | 2249 | 0.394 | 0.2237 | Yes |
| 24 | Fbln2 | 2410 | 0.381 | 0.2251 | Yes |
| 25 | Col1a1 | 2477 | 0.377 | 0.2312 | Yes |
| 26 | Col12a1 | 2781 | 0.357 | 0.2249 | Yes |
| 27 | Cald1 | 2832 | 0.355 | 0.2312 | Yes |
| 28 | Pcolce2 | 3030 | 0.340 | 0.2297 | Yes |
| 29 | Wipf1 | 3103 | 0.339 | 0.2346 | Yes |
| 30 | Gpc1 | 3114 | 0.338 | 0.2425 | Yes |
| 31 | Gm21451 | 3362 | 0.322 | 0.2381 | Yes |
| 32 | Slc6a8 | 3378 | 0.321 | 0.2453 | Yes |
| 33 | Dcn | 3438 | 0.316 | 0.2502 | Yes |
| 34 | Mmp2 | 3859 | 0.284 | 0.2362 | No |
| 35 | Fzd8 | 3919 | 0.279 | 0.2401 | No |
| 36 | Tnfrsf12a | 3959 | 0.275 | 0.2450 | No |
| 37 | Itgav | 4486 | 0.243 | 0.2246 | No |
| 38 | Flna | 5084 | 0.210 | 0.1998 | No |
| 39 | Pvr | 5148 | 0.206 | 0.2018 | No |
| 40 | Notch2 | 5185 | 0.204 | 0.2051 | No |
| 41 | Col1a2 | 5315 | 0.200 | 0.2036 | No |
| 42 | Mest | 5510 | 0.193 | 0.1986 | No |
| 43 | Eno2 | 5556 | 0.191 | 0.2011 | No |
| 44 | Nid2 | 5686 | 0.189 | 0.1993 | No |
| 45 | Tnfaip3 | 5956 | 0.173 | 0.1901 | No |
| 46 | Tgfbi | 5957 | 0.173 | 0.1944 | No |
| 47 | Jun | 6045 | 0.170 | 0.1943 | No |
| 48 | Pcolce | 6186 | 0.168 | 0.1914 | No |
| 49 | Sparc | 6210 | 0.166 | 0.1944 | No |
| 50 | Plod3 | 6315 | 0.161 | 0.1932 | No |
| 51 | Gem | 6336 | 0.160 | 0.1962 | No |
| 52 | Gadd45a | 6370 | 0.158 | 0.1984 | No |
| 53 | Postn | 6489 | 0.152 | 0.1963 | No |
| 54 | Copa | 6545 | 0.149 | 0.1972 | No |
| 55 | Mmp14 | 6563 | 0.148 | 0.2001 | No |
| 56 | Timp3 | 6600 | 0.146 | 0.2019 | No |
| 57 | Sdc1 | 6848 | 0.135 | 0.1928 | No |
| 58 | Lama1 | 6939 | 0.132 | 0.1916 | No |
| 59 | Tgm2 | 7259 | 0.116 | 0.1785 | No |
| 60 | Sgcd | 7355 | 0.112 | 0.1765 | No |
| 61 | Col11a1 | 7580 | 0.102 | 0.1678 | No |
| 62 | Col7a1 | 7661 | 0.099 | 0.1662 | No |
| 63 | Lama3 | 7718 | 0.096 | 0.1658 | No |
| 64 | Spp1 | 7915 | 0.088 | 0.1581 | No |
| 65 | Calu | 7957 | 0.087 | 0.1582 | No |
| 66 | Nt5e | 8212 | 0.077 | 0.1473 | No |
| 67 | Thbs1 | 8736 | 0.055 | 0.1224 | No |
| 68 | Rhob | 8920 | 0.048 | 0.1144 | No |
| 69 | Fap | 9122 | 0.040 | 0.1053 | No |
| 70 | Lama2 | 9183 | 0.038 | 0.1032 | No |
| 71 | Tgfb1 | 9241 | 0.035 | 0.1012 | No |
| 72 | Matn2 | 9445 | 0.027 | 0.0917 | No |
| 73 | Il15 | 9513 | 0.024 | 0.0889 | No |
| 74 | Slit3 | 9664 | 0.018 | 0.0818 | No |
| 75 | Fbln5 | 10375 | -0.003 | 0.0462 | No |
| 76 | Itga5 | 10640 | -0.012 | 0.0332 | No |
| 77 | Itgb3 | 10934 | -0.022 | 0.0190 | No |
| 78 | Bgn | 10991 | -0.025 | 0.0168 | No |
| 79 | Cadm1 | 11076 | -0.028 | 0.0133 | No |
| 80 | Fas | 11107 | -0.030 | 0.0125 | No |
| 81 | Tpm4 | 11176 | -0.032 | 0.0099 | No |
| 82 | Tagln | 11270 | -0.036 | 0.0061 | No |
| 83 | Ntm | 11439 | -0.043 | -0.0013 | No |
| 84 | Pmepa1 | 11639 | -0.051 | -0.0100 | No |
| 85 | Fn1 | 11719 | -0.054 | -0.0126 | No |
| 86 | Fbln1 | 11822 | -0.058 | -0.0163 | No |
| 87 | Capg | 11875 | -0.061 | -0.0174 | No |
| 88 | Vegfc | 11944 | -0.064 | -0.0192 | No |
| 89 | Cd44 | 12079 | -0.069 | -0.0242 | No |
| 90 | Lrp1 | 12207 | -0.075 | -0.0288 | No |
| 91 | Qsox1 | 12466 | -0.087 | -0.0396 | No |
| 92 | Plod1 | 12724 | -0.099 | -0.0500 | No |
| 93 | Magee1 | 13024 | -0.109 | -0.0624 | No |
| 94 | Plaur | 13226 | -0.119 | -0.0695 | No |
| 95 | Pfn2 | 13301 | -0.122 | -0.0702 | No |
| 96 | Itga2 | 13383 | -0.125 | -0.0711 | No |
| 97 | Serpine2 | 13393 | -0.125 | -0.0685 | No |
| 98 | Comp | 13444 | -0.128 | -0.0678 | No |
| 99 | Anpep | 13753 | -0.141 | -0.0798 | No |
| 100 | Gja1 | 13797 | -0.142 | -0.0784 | No |
| 101 | Glipr1 | 14064 | -0.154 | -0.0879 | No |
| 102 | Abi3bp | 14081 | -0.155 | -0.0849 | No |
| 103 | Pdgfrb | 14303 | -0.164 | -0.0919 | No |
| 104 | Tnc | 14433 | -0.170 | -0.0942 | No |
| 105 | Dst | 14476 | -0.172 | -0.0920 | No |
| 106 | Sntb1 | 14518 | -0.174 | -0.0897 | No |
| 107 | Sgcb | 14696 | -0.183 | -0.0941 | No |
| 108 | Itgb1 | 14925 | -0.193 | -0.1007 | No |
| 109 | Myl9 | 14984 | -0.196 | -0.0988 | No |
| 110 | Col4a2 | 15442 | -0.218 | -0.1163 | No |
| 111 | Mcm7 | 16224 | -0.259 | -0.1492 | No |
| 112 | Tpm1 | 16344 | -0.263 | -0.1486 | No |
| 113 | Id2 | 16403 | -0.266 | -0.1449 | No |
| 114 | Gadd45b | 16482 | -0.272 | -0.1420 | No |
| 115 | Cdh2 | 16814 | -0.291 | -0.1514 | No |
| 116 | Fstl1 | 16869 | -0.295 | -0.1468 | No |
| 117 | P3h1 | 16883 | -0.295 | -0.1401 | No |
| 118 | Loxl1 | 17028 | -0.304 | -0.1398 | No |
| 119 | Tfpi2 | 17245 | -0.319 | -0.1427 | No |
| 120 | Dpysl3 | 17419 | -0.329 | -0.1432 | No |
| 121 | Bmp1 | 17534 | -0.336 | -0.1405 | No |
| 122 | Pdlim4 | 17638 | -0.343 | -0.1372 | No |
| 123 | Cdh6 | 17658 | -0.344 | -0.1296 | No |
| 124 | Plod2 | 17779 | -0.354 | -0.1268 | No |
| 125 | Col4a1 | 18084 | -0.381 | -0.1326 | No |
| 126 | Col5a2 | 18139 | -0.385 | -0.1257 | No |
| 127 | Cdh11 | 18507 | -0.419 | -0.1337 | No |
| 128 | Fgf2 | 18566 | -0.426 | -0.1260 | No |
| 129 | Colgalt1 | 18638 | -0.434 | -0.1188 | No |
| 130 | Thy1 | 18642 | -0.435 | -0.1081 | No |
| 131 | Sfrp4 | 18913 | -0.472 | -0.1099 | No |
| 132 | Basp1 | 19058 | -0.496 | -0.1048 | No |
| 133 | Tgfbr3 | 19107 | -0.505 | -0.0947 | No |
| 134 | Fbn1 | 19155 | -0.515 | -0.0842 | No |
| 135 | Spock1 | 19290 | -0.539 | -0.0775 | No |
| 136 | Igfbp4 | 19335 | -0.550 | -0.0660 | No |
| 137 | Emp3 | 19385 | -0.563 | -0.0545 | No |
| 138 | Vim | 19425 | -0.574 | -0.0421 | No |
| 139 | Ppib | 19603 | -0.637 | -0.0352 | No |
| 140 | Fbn2 | 19636 | -0.651 | -0.0206 | No |
| 141 | Col16a1 | 19723 | -0.686 | -0.0078 | No |
| 142 | Lgals1 | 19975 | -0.923 | 0.0026 | No |