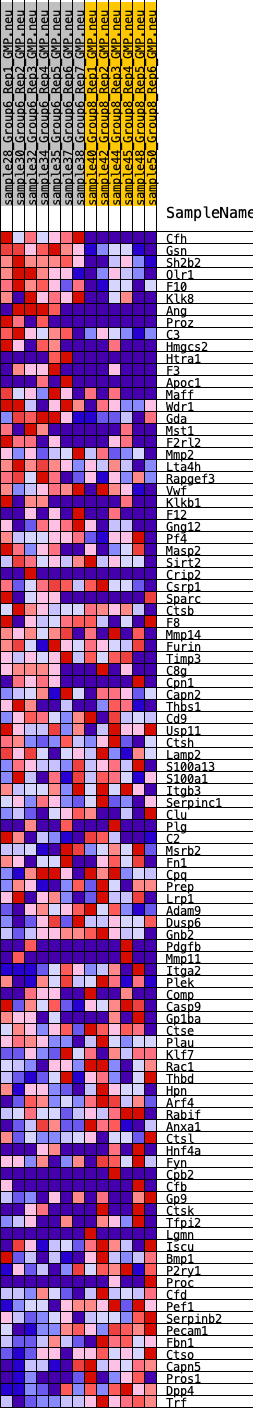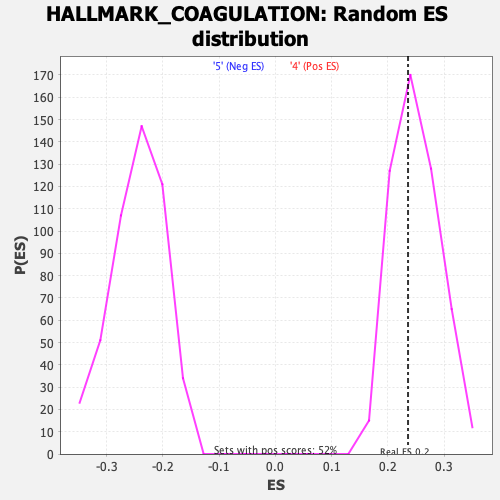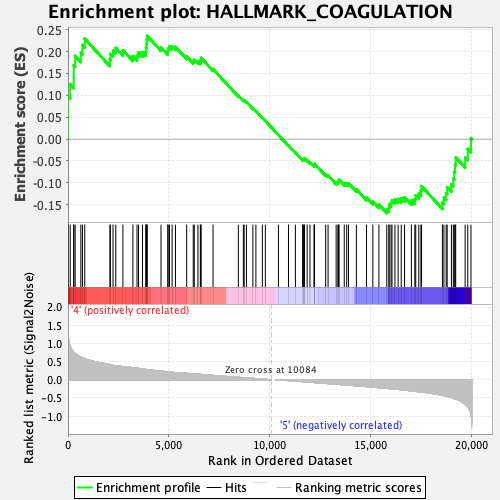
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group6_versus_Group8.GMP.neu_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | GMP.neu_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | 0.23571007 |
| Normalized Enrichment Score (NES) | 0.94459957 |
| Nominal p-value | 0.5628627 |
| FDR q-value | 0.7385882 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cfh | 4 | 1.496 | 0.0518 | Yes |
| 2 | Gsn | 9 | 1.326 | 0.0977 | Yes |
| 3 | Sh2b2 | 109 | 0.926 | 0.1249 | Yes |
| 4 | Olr1 | 285 | 0.754 | 0.1424 | Yes |
| 5 | F10 | 286 | 0.754 | 0.1686 | Yes |
| 6 | Klk8 | 355 | 0.715 | 0.1900 | Yes |
| 7 | Ang | 637 | 0.625 | 0.1977 | Yes |
| 8 | Proz | 717 | 0.605 | 0.2147 | Yes |
| 9 | C3 | 830 | 0.578 | 0.2292 | Yes |
| 10 | Hmgcs2 | 2079 | 0.417 | 0.1811 | Yes |
| 11 | Htra1 | 2100 | 0.415 | 0.1945 | Yes |
| 12 | F3 | 2233 | 0.396 | 0.2017 | Yes |
| 13 | Apoc1 | 2370 | 0.384 | 0.2082 | Yes |
| 14 | Maff | 2723 | 0.361 | 0.2031 | Yes |
| 15 | Wdr1 | 3219 | 0.329 | 0.1897 | Yes |
| 16 | Gda | 3423 | 0.317 | 0.1905 | Yes |
| 17 | Mst1 | 3495 | 0.314 | 0.1979 | Yes |
| 18 | F2rl2 | 3695 | 0.298 | 0.1983 | Yes |
| 19 | Mmp2 | 3859 | 0.284 | 0.2000 | Yes |
| 20 | Lta4h | 3873 | 0.283 | 0.2091 | Yes |
| 21 | Rapgef3 | 3898 | 0.281 | 0.2177 | Yes |
| 22 | Vwf | 3906 | 0.280 | 0.2271 | Yes |
| 23 | Klkb1 | 3928 | 0.278 | 0.2357 | Yes |
| 24 | F12 | 4612 | 0.237 | 0.2097 | No |
| 25 | Gng12 | 4948 | 0.218 | 0.2004 | No |
| 26 | Pf4 | 4967 | 0.217 | 0.2071 | No |
| 27 | Masp2 | 5031 | 0.213 | 0.2113 | No |
| 28 | Sirt2 | 5164 | 0.205 | 0.2118 | No |
| 29 | Crip2 | 5329 | 0.199 | 0.2105 | No |
| 30 | Csrp1 | 5886 | 0.177 | 0.1888 | No |
| 31 | Sparc | 6210 | 0.166 | 0.1783 | No |
| 32 | Ctsb | 6263 | 0.164 | 0.1814 | No |
| 33 | F8 | 6443 | 0.154 | 0.1778 | No |
| 34 | Mmp14 | 6563 | 0.148 | 0.1770 | No |
| 35 | Furin | 6583 | 0.147 | 0.1811 | No |
| 36 | Timp3 | 6600 | 0.146 | 0.1854 | No |
| 37 | C8g | 7193 | 0.120 | 0.1599 | No |
| 38 | Cpn1 | 8448 | 0.067 | 0.0993 | No |
| 39 | Capn2 | 8696 | 0.057 | 0.0888 | No |
| 40 | Thbs1 | 8736 | 0.055 | 0.0888 | No |
| 41 | Cd9 | 8849 | 0.051 | 0.0850 | No |
| 42 | Usp11 | 9169 | 0.038 | 0.0703 | No |
| 43 | Ctsh | 9318 | 0.032 | 0.0640 | No |
| 44 | Lamp2 | 9639 | 0.019 | 0.0486 | No |
| 45 | S100a13 | 9787 | 0.013 | 0.0416 | No |
| 46 | S100a1 | 10436 | -0.005 | 0.0093 | No |
| 47 | Itgb3 | 10934 | -0.022 | -0.0149 | No |
| 48 | Serpinc1 | 11277 | -0.036 | -0.0308 | No |
| 49 | Clu | 11633 | -0.050 | -0.0468 | No |
| 50 | Plg | 11693 | -0.053 | -0.0480 | No |
| 51 | C2 | 11696 | -0.053 | -0.0462 | No |
| 52 | Msrb2 | 11717 | -0.054 | -0.0453 | No |
| 53 | Fn1 | 11719 | -0.054 | -0.0435 | No |
| 54 | Cpq | 11860 | -0.060 | -0.0484 | No |
| 55 | Prep | 12001 | -0.066 | -0.0532 | No |
| 56 | Lrp1 | 12207 | -0.075 | -0.0608 | No |
| 57 | Adam9 | 12212 | -0.076 | -0.0584 | No |
| 58 | Dusp6 | 12226 | -0.076 | -0.0564 | No |
| 59 | Gnb2 | 12780 | -0.101 | -0.0806 | No |
| 60 | Pdgfb | 12898 | -0.105 | -0.0829 | No |
| 61 | Mmp11 | 13298 | -0.122 | -0.0986 | No |
| 62 | Itga2 | 13383 | -0.125 | -0.0985 | No |
| 63 | Plek | 13418 | -0.127 | -0.0958 | No |
| 64 | Comp | 13444 | -0.128 | -0.0926 | No |
| 65 | Casp9 | 13700 | -0.139 | -0.1006 | No |
| 66 | Gp1ba | 13821 | -0.143 | -0.1016 | No |
| 67 | Ctse | 13910 | -0.147 | -0.1009 | No |
| 68 | Plau | 14301 | -0.163 | -0.1148 | No |
| 69 | Klf7 | 14804 | -0.187 | -0.1335 | No |
| 70 | Rac1 | 15118 | -0.203 | -0.1422 | No |
| 71 | Thbd | 15419 | -0.217 | -0.1497 | No |
| 72 | Hpn | 15809 | -0.236 | -0.1610 | No |
| 73 | Arf4 | 15915 | -0.241 | -0.1579 | No |
| 74 | Rabif | 15925 | -0.242 | -0.1499 | No |
| 75 | Anxa1 | 16020 | -0.247 | -0.1461 | No |
| 76 | Ctsl | 16068 | -0.251 | -0.1397 | No |
| 77 | Hnf4a | 16213 | -0.258 | -0.1379 | No |
| 78 | Fyn | 16371 | -0.265 | -0.1366 | No |
| 79 | Cpb2 | 16526 | -0.274 | -0.1348 | No |
| 80 | Cfb | 16689 | -0.282 | -0.1332 | No |
| 81 | Gp9 | 17030 | -0.304 | -0.1396 | No |
| 82 | Ctsk | 17201 | -0.317 | -0.1372 | No |
| 83 | Tfpi2 | 17245 | -0.319 | -0.1282 | No |
| 84 | Lgmn | 17397 | -0.328 | -0.1244 | No |
| 85 | Iscu | 17502 | -0.335 | -0.1180 | No |
| 86 | Bmp1 | 17534 | -0.336 | -0.1078 | No |
| 87 | P2ry1 | 18569 | -0.426 | -0.1449 | No |
| 88 | Proc | 18643 | -0.435 | -0.1335 | No |
| 89 | Cfd | 18754 | -0.450 | -0.1233 | No |
| 90 | Pef1 | 18804 | -0.457 | -0.1099 | No |
| 91 | Serpinb2 | 19013 | -0.486 | -0.1034 | No |
| 92 | Pecam1 | 19109 | -0.505 | -0.0906 | No |
| 93 | Fbn1 | 19155 | -0.515 | -0.0750 | No |
| 94 | Ctso | 19195 | -0.523 | -0.0588 | No |
| 95 | Capn5 | 19226 | -0.529 | -0.0419 | No |
| 96 | Pros1 | 19695 | -0.674 | -0.0420 | No |
| 97 | Dpp4 | 19826 | -0.747 | -0.0225 | No |
| 98 | Trf | 19984 | -0.935 | 0.0021 | No |