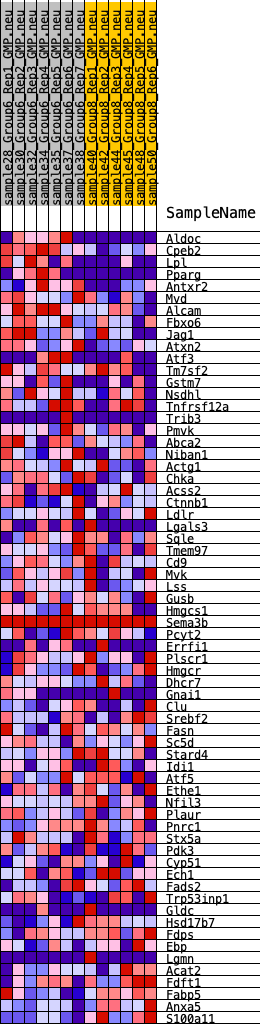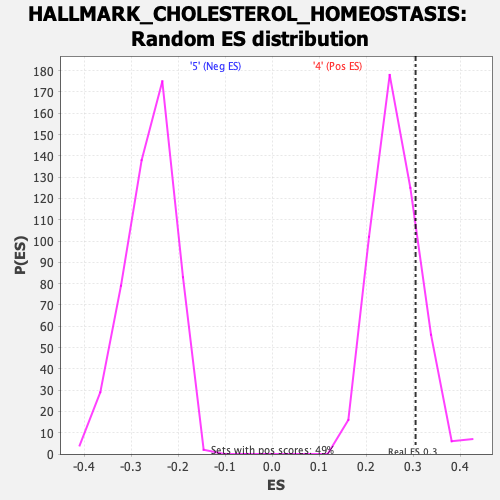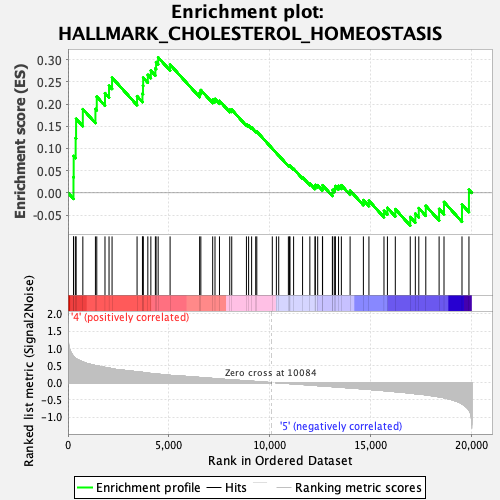
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group6_versus_Group8.GMP.neu_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | GMP.neu_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | 0.30499315 |
| Normalized Enrichment Score (NES) | 1.1632106 |
| Nominal p-value | 0.17142858 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.949 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Aldoc | 272 | 0.764 | 0.0351 | Yes |
| 2 | Cpeb2 | 282 | 0.757 | 0.0830 | Yes |
| 3 | Lpl | 379 | 0.703 | 0.1231 | Yes |
| 4 | Pparg | 401 | 0.699 | 0.1668 | Yes |
| 5 | Antxr2 | 736 | 0.601 | 0.1884 | Yes |
| 6 | Mvd | 1360 | 0.495 | 0.1889 | Yes |
| 7 | Alcam | 1427 | 0.485 | 0.2165 | Yes |
| 8 | Fbxo6 | 1834 | 0.442 | 0.2244 | Yes |
| 9 | Jag1 | 2035 | 0.421 | 0.2413 | Yes |
| 10 | Atxn2 | 2185 | 0.402 | 0.2595 | Yes |
| 11 | Atf3 | 3425 | 0.317 | 0.2177 | Yes |
| 12 | Tm7sf2 | 3692 | 0.299 | 0.2234 | Yes |
| 13 | Gstm7 | 3719 | 0.297 | 0.2411 | Yes |
| 14 | Nsdhl | 3728 | 0.296 | 0.2596 | Yes |
| 15 | Tnfrsf12a | 3959 | 0.275 | 0.2657 | Yes |
| 16 | Trib3 | 4109 | 0.266 | 0.2752 | Yes |
| 17 | Pmvk | 4333 | 0.253 | 0.2802 | Yes |
| 18 | Abca2 | 4368 | 0.250 | 0.2945 | Yes |
| 19 | Niban1 | 4471 | 0.244 | 0.3050 | Yes |
| 20 | Actg1 | 5063 | 0.211 | 0.2888 | No |
| 21 | Chka | 6528 | 0.150 | 0.2251 | No |
| 22 | Acss2 | 6588 | 0.147 | 0.2315 | No |
| 23 | Ctnnb1 | 7175 | 0.121 | 0.2098 | No |
| 24 | Ldlr | 7290 | 0.115 | 0.2115 | No |
| 25 | Lgals3 | 7512 | 0.105 | 0.2072 | No |
| 26 | Sqle | 8021 | 0.084 | 0.1871 | No |
| 27 | Tmem97 | 8120 | 0.080 | 0.1873 | No |
| 28 | Cd9 | 8849 | 0.051 | 0.1541 | No |
| 29 | Mvk | 8957 | 0.047 | 0.1518 | No |
| 30 | Lss | 9106 | 0.040 | 0.1469 | No |
| 31 | Gusb | 9309 | 0.032 | 0.1389 | No |
| 32 | Hmgcs1 | 9363 | 0.030 | 0.1381 | No |
| 33 | Sema3b | 10132 | 0.000 | 0.0996 | No |
| 34 | Pcyt2 | 10334 | -0.001 | 0.0896 | No |
| 35 | Errfi1 | 10450 | -0.006 | 0.0842 | No |
| 36 | Plscr1 | 10929 | -0.022 | 0.0617 | No |
| 37 | Hmgcr | 10982 | -0.024 | 0.0607 | No |
| 38 | Dhcr7 | 11009 | -0.025 | 0.0610 | No |
| 39 | Gnai1 | 11188 | -0.032 | 0.0541 | No |
| 40 | Clu | 11633 | -0.050 | 0.0351 | No |
| 41 | Srebf2 | 11994 | -0.066 | 0.0213 | No |
| 42 | Fasn | 12250 | -0.077 | 0.0135 | No |
| 43 | Sc5d | 12264 | -0.078 | 0.0178 | No |
| 44 | Stard4 | 12383 | -0.083 | 0.0172 | No |
| 45 | Idi1 | 12619 | -0.094 | 0.0114 | No |
| 46 | Atf5 | 12626 | -0.094 | 0.0171 | No |
| 47 | Ethe1 | 13111 | -0.113 | 0.0001 | No |
| 48 | Nfil3 | 13130 | -0.114 | 0.0065 | No |
| 49 | Plaur | 13226 | -0.119 | 0.0094 | No |
| 50 | Pnrc1 | 13261 | -0.120 | 0.0154 | No |
| 51 | Stx5a | 13417 | -0.127 | 0.0157 | No |
| 52 | Pdk3 | 13557 | -0.132 | 0.0172 | No |
| 53 | Cyp51 | 13990 | -0.150 | 0.0051 | No |
| 54 | Ech1 | 14649 | -0.180 | -0.0164 | No |
| 55 | Fads2 | 14924 | -0.193 | -0.0177 | No |
| 56 | Trp53inp1 | 15670 | -0.229 | -0.0404 | No |
| 57 | Gldc | 15841 | -0.238 | -0.0337 | No |
| 58 | Hsd17b7 | 16232 | -0.259 | -0.0367 | No |
| 59 | Fdps | 16973 | -0.301 | -0.0545 | No |
| 60 | Ebp | 17225 | -0.318 | -0.0468 | No |
| 61 | Lgmn | 17397 | -0.328 | -0.0344 | No |
| 62 | Acat2 | 17744 | -0.351 | -0.0293 | No |
| 63 | Fdft1 | 18404 | -0.408 | -0.0362 | No |
| 64 | Fabp5 | 18647 | -0.435 | -0.0206 | No |
| 65 | Anxa5 | 19539 | -0.611 | -0.0262 | No |
| 66 | S100a11 | 19884 | -0.791 | 0.0071 | No |