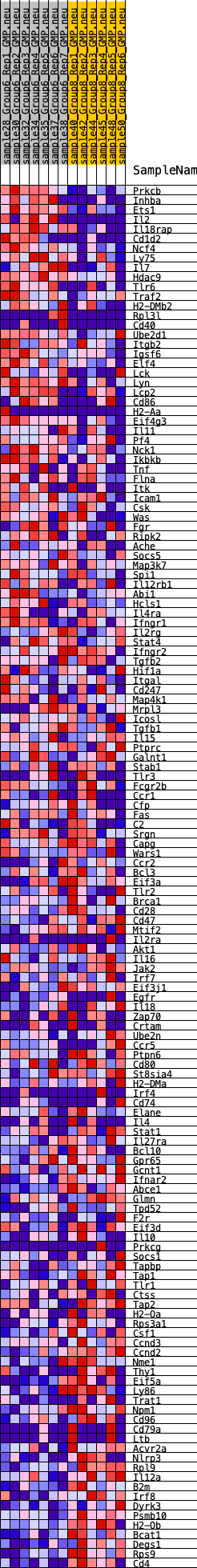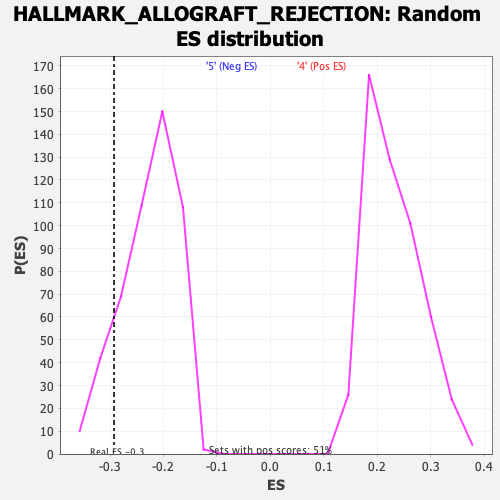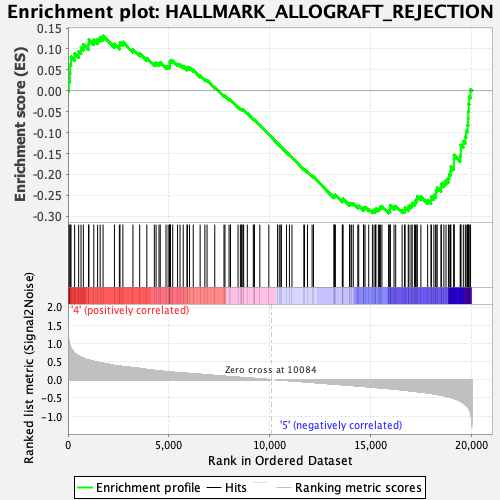
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group6_versus_Group8.GMP.neu_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | GMP.neu_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_ALLOGRAFT_REJECTION |
| Enrichment Score (ES) | -0.29251605 |
| Normalized Enrichment Score (NES) | -1.2895321 |
| Nominal p-value | 0.1122449 |
| FDR q-value | 0.20231776 |
| FWER p-Value | 0.77 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Prkcb | 39 | 1.086 | 0.0232 | No |
| 2 | Inhba | 93 | 0.946 | 0.0424 | No |
| 3 | Ets1 | 107 | 0.927 | 0.0632 | No |
| 4 | Il2 | 150 | 0.871 | 0.0812 | No |
| 5 | Il18rap | 328 | 0.728 | 0.0892 | No |
| 6 | Cd1d2 | 528 | 0.654 | 0.0943 | No |
| 7 | Ncf4 | 646 | 0.622 | 0.1028 | No |
| 8 | Ly75 | 761 | 0.596 | 0.1109 | No |
| 9 | Il7 | 1019 | 0.543 | 0.1105 | No |
| 10 | Hdac9 | 1037 | 0.543 | 0.1222 | No |
| 11 | Tlr6 | 1279 | 0.505 | 0.1218 | No |
| 12 | Traf2 | 1469 | 0.482 | 0.1235 | No |
| 13 | H2-DMb2 | 1595 | 0.469 | 0.1280 | No |
| 14 | Rpl3l | 1738 | 0.455 | 0.1314 | No |
| 15 | Cd40 | 2303 | 0.389 | 0.1120 | No |
| 16 | Ube2d1 | 2547 | 0.373 | 0.1084 | No |
| 17 | Itgb2 | 2578 | 0.371 | 0.1155 | No |
| 18 | Igsf6 | 2721 | 0.361 | 0.1167 | No |
| 19 | Elf4 | 3220 | 0.329 | 0.0993 | No |
| 20 | Lck | 3555 | 0.308 | 0.0896 | No |
| 21 | Lyn | 3910 | 0.280 | 0.0783 | No |
| 22 | Lcp2 | 4282 | 0.255 | 0.0656 | No |
| 23 | Cd86 | 4369 | 0.250 | 0.0670 | No |
| 24 | H2-Aa | 4514 | 0.241 | 0.0654 | No |
| 25 | Eif4g3 | 4574 | 0.239 | 0.0679 | No |
| 26 | Il11 | 4860 | 0.223 | 0.0588 | No |
| 27 | Pf4 | 4967 | 0.217 | 0.0584 | No |
| 28 | Nck1 | 5044 | 0.212 | 0.0595 | No |
| 29 | Ikbkb | 5048 | 0.212 | 0.0643 | No |
| 30 | Tnf | 5052 | 0.211 | 0.0690 | No |
| 31 | Flna | 5084 | 0.210 | 0.0723 | No |
| 32 | Itk | 5193 | 0.203 | 0.0716 | No |
| 33 | Icam1 | 5433 | 0.197 | 0.0641 | No |
| 34 | Csk | 5555 | 0.191 | 0.0624 | No |
| 35 | Was | 5714 | 0.187 | 0.0588 | No |
| 36 | Fgr | 5901 | 0.176 | 0.0535 | No |
| 37 | Ripk2 | 5920 | 0.175 | 0.0567 | No |
| 38 | Ache | 6026 | 0.171 | 0.0554 | No |
| 39 | Socs5 | 6212 | 0.166 | 0.0499 | No |
| 40 | Map3k7 | 6553 | 0.149 | 0.0362 | No |
| 41 | Spi1 | 6788 | 0.138 | 0.0277 | No |
| 42 | Il12rb1 | 6895 | 0.133 | 0.0254 | No |
| 43 | Abi1 | 7278 | 0.116 | 0.0089 | No |
| 44 | Hcls1 | 7736 | 0.095 | -0.0119 | No |
| 45 | Il4ra | 7784 | 0.093 | -0.0121 | No |
| 46 | Ifngr1 | 7987 | 0.086 | -0.0203 | No |
| 47 | Il2rg | 8058 | 0.083 | -0.0219 | No |
| 48 | Stat4 | 8433 | 0.067 | -0.0391 | No |
| 49 | Ifngr2 | 8549 | 0.063 | -0.0435 | No |
| 50 | Tgfb2 | 8594 | 0.061 | -0.0443 | No |
| 51 | Hif1a | 8623 | 0.060 | -0.0443 | No |
| 52 | Itgal | 8678 | 0.058 | -0.0457 | No |
| 53 | Cd247 | 8712 | 0.056 | -0.0460 | No |
| 54 | Map4k1 | 8893 | 0.049 | -0.0540 | No |
| 55 | Mrpl3 | 9190 | 0.038 | -0.0680 | No |
| 56 | Icosl | 9229 | 0.036 | -0.0691 | No |
| 57 | Tgfb1 | 9241 | 0.035 | -0.0688 | No |
| 58 | Il15 | 9513 | 0.024 | -0.0819 | No |
| 59 | Ptprc | 9960 | 0.005 | -0.1042 | No |
| 60 | Galnt1 | 10393 | -0.004 | -0.1258 | No |
| 61 | Stab1 | 10477 | -0.007 | -0.1299 | No |
| 62 | Tlr3 | 10557 | -0.009 | -0.1336 | No |
| 63 | Fcgr2b | 10582 | -0.010 | -0.1346 | No |
| 64 | Ccr1 | 10835 | -0.018 | -0.1468 | No |
| 65 | Cfp | 10981 | -0.024 | -0.1536 | No |
| 66 | Fas | 11107 | -0.030 | -0.1592 | No |
| 67 | C2 | 11696 | -0.053 | -0.1875 | No |
| 68 | Srgn | 11730 | -0.054 | -0.1879 | No |
| 69 | Capg | 11875 | -0.061 | -0.1938 | No |
| 70 | Wars1 | 12106 | -0.071 | -0.2037 | No |
| 71 | Ccr2 | 12174 | -0.074 | -0.2053 | No |
| 72 | Bcl3 | 13184 | -0.117 | -0.2534 | No |
| 73 | Eif3a | 13217 | -0.118 | -0.2522 | No |
| 74 | Tlr2 | 13219 | -0.119 | -0.2495 | No |
| 75 | Brca1 | 13260 | -0.120 | -0.2488 | No |
| 76 | Cd28 | 13603 | -0.135 | -0.2629 | No |
| 77 | Cd47 | 13623 | -0.136 | -0.2607 | No |
| 78 | Mtif2 | 13638 | -0.136 | -0.2582 | No |
| 79 | Il2ra | 13961 | -0.149 | -0.2710 | No |
| 80 | Akt1 | 13985 | -0.150 | -0.2686 | No |
| 81 | Il16 | 14066 | -0.154 | -0.2691 | No |
| 82 | Jak2 | 14148 | -0.158 | -0.2695 | No |
| 83 | Irf7 | 14372 | -0.167 | -0.2769 | No |
| 84 | Eif3j1 | 14417 | -0.169 | -0.2752 | No |
| 85 | Egfr | 14654 | -0.180 | -0.2829 | No |
| 86 | Il18 | 14671 | -0.181 | -0.2795 | No |
| 87 | Zap70 | 14731 | -0.184 | -0.2782 | No |
| 88 | Crtam | 14914 | -0.193 | -0.2829 | No |
| 89 | Ube2n | 15096 | -0.202 | -0.2873 | Yes |
| 90 | Ccr5 | 15178 | -0.205 | -0.2866 | Yes |
| 91 | Ptpn6 | 15249 | -0.208 | -0.2853 | Yes |
| 92 | Cd80 | 15271 | -0.209 | -0.2816 | Yes |
| 93 | St8sia4 | 15393 | -0.216 | -0.2826 | Yes |
| 94 | H2-DMa | 15445 | -0.218 | -0.2802 | Yes |
| 95 | Irf4 | 15491 | -0.221 | -0.2773 | Yes |
| 96 | Cd74 | 15567 | -0.224 | -0.2759 | Yes |
| 97 | Elane | 15898 | -0.240 | -0.2870 | Yes |
| 98 | Il4 | 15970 | -0.244 | -0.2849 | Yes |
| 99 | Stat1 | 15971 | -0.244 | -0.2792 | Yes |
| 100 | Il27ra | 15974 | -0.244 | -0.2737 | Yes |
| 101 | Bcl10 | 16168 | -0.255 | -0.2775 | Yes |
| 102 | Gpr65 | 16251 | -0.260 | -0.2756 | Yes |
| 103 | Gcnt1 | 16570 | -0.274 | -0.2852 | Yes |
| 104 | Ifnar2 | 16691 | -0.282 | -0.2847 | Yes |
| 105 | Abce1 | 16718 | -0.285 | -0.2794 | Yes |
| 106 | Glmn | 16874 | -0.295 | -0.2804 | Yes |
| 107 | Tpd52 | 16907 | -0.297 | -0.2752 | Yes |
| 108 | F2r | 17012 | -0.303 | -0.2734 | Yes |
| 109 | Eif3d | 17069 | -0.308 | -0.2691 | Yes |
| 110 | Il10 | 17181 | -0.315 | -0.2673 | Yes |
| 111 | Prkcg | 17232 | -0.319 | -0.2625 | Yes |
| 112 | Socs1 | 17299 | -0.323 | -0.2583 | Yes |
| 113 | Tapbp | 17321 | -0.324 | -0.2519 | Yes |
| 114 | Tap1 | 17500 | -0.335 | -0.2531 | Yes |
| 115 | Tlr1 | 17836 | -0.359 | -0.2616 | Yes |
| 116 | Ctss | 18002 | -0.373 | -0.2613 | Yes |
| 117 | Tap2 | 18019 | -0.374 | -0.2534 | Yes |
| 118 | H2-Oa | 18146 | -0.386 | -0.2508 | Yes |
| 119 | Rps3a1 | 18238 | -0.393 | -0.2463 | Yes |
| 120 | Csf1 | 18247 | -0.394 | -0.2376 | Yes |
| 121 | Ccnd3 | 18310 | -0.400 | -0.2315 | Yes |
| 122 | Ccnd2 | 18500 | -0.419 | -0.2313 | Yes |
| 123 | Nme1 | 18513 | -0.420 | -0.2222 | Yes |
| 124 | Thy1 | 18642 | -0.435 | -0.2185 | Yes |
| 125 | Eif5a | 18747 | -0.448 | -0.2134 | Yes |
| 126 | Ly86 | 18867 | -0.467 | -0.2086 | Yes |
| 127 | Trat1 | 18906 | -0.471 | -0.1996 | Yes |
| 128 | Npm1 | 18973 | -0.481 | -0.1918 | Yes |
| 129 | Cd96 | 18986 | -0.483 | -0.1812 | Yes |
| 130 | Cd79a | 19138 | -0.512 | -0.1769 | Yes |
| 131 | Ltb | 19139 | -0.512 | -0.1651 | Yes |
| 132 | Acvr2a | 19144 | -0.513 | -0.1534 | Yes |
| 133 | Nlrp3 | 19450 | -0.581 | -0.1553 | Yes |
| 134 | Rpl9 | 19480 | -0.593 | -0.1430 | Yes |
| 135 | Il12a | 19481 | -0.594 | -0.1293 | Yes |
| 136 | B2m | 19605 | -0.640 | -0.1207 | Yes |
| 137 | Irf8 | 19704 | -0.679 | -0.1099 | Yes |
| 138 | Dyrk3 | 19745 | -0.702 | -0.0957 | Yes |
| 139 | Psmb10 | 19813 | -0.734 | -0.0821 | Yes |
| 140 | H2-Ob | 19844 | -0.759 | -0.0660 | Yes |
| 141 | Bcat1 | 19853 | -0.764 | -0.0487 | Yes |
| 142 | Degs1 | 19874 | -0.785 | -0.0316 | Yes |
| 143 | Rps9 | 19895 | -0.801 | -0.0140 | Yes |
| 144 | Cd4 | 19963 | -0.889 | 0.0032 | Yes |