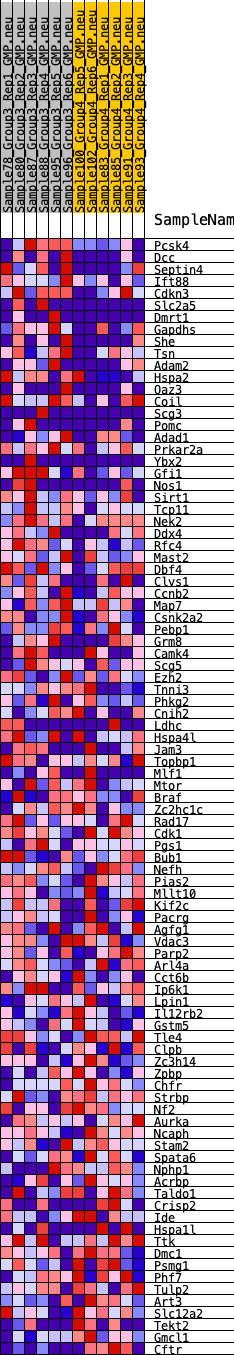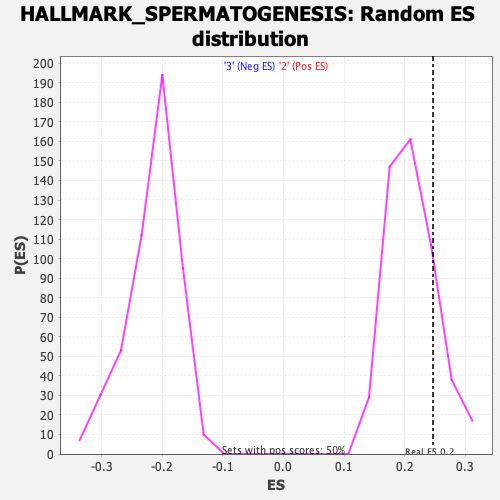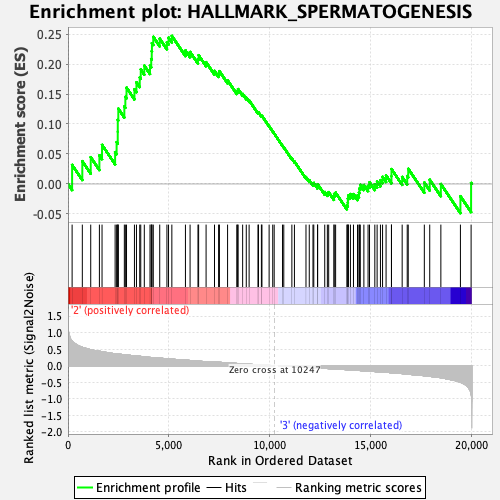
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group3_versus_Group4.GMP.neu_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.2469375 |
| Normalized Enrichment Score (NES) | 1.1717507 |
| Nominal p-value | 0.15631263 |
| FDR q-value | 0.8643101 |
| FWER p-Value | 0.919 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pcsk4 | 204 | 0.744 | 0.0319 | Yes |
| 2 | Dcc | 710 | 0.557 | 0.0380 | Yes |
| 3 | Septin4 | 1127 | 0.486 | 0.0447 | Yes |
| 4 | Ift88 | 1557 | 0.435 | 0.0478 | Yes |
| 5 | Cdkn3 | 1688 | 0.424 | 0.0652 | Yes |
| 6 | Slc2a5 | 2337 | 0.361 | 0.0532 | Yes |
| 7 | Dmrt1 | 2411 | 0.357 | 0.0697 | Yes |
| 8 | Gapdhs | 2467 | 0.355 | 0.0870 | Yes |
| 9 | She | 2469 | 0.354 | 0.1070 | Yes |
| 10 | Tsn | 2495 | 0.352 | 0.1257 | Yes |
| 11 | Adam2 | 2794 | 0.330 | 0.1294 | Yes |
| 12 | Hspa2 | 2844 | 0.325 | 0.1454 | Yes |
| 13 | Oaz3 | 2902 | 0.320 | 0.1606 | Yes |
| 14 | Coil | 3295 | 0.300 | 0.1580 | Yes |
| 15 | Scg3 | 3395 | 0.293 | 0.1696 | Yes |
| 16 | Pomc | 3558 | 0.285 | 0.1776 | Yes |
| 17 | Adad1 | 3608 | 0.281 | 0.1910 | Yes |
| 18 | Prkar2a | 3784 | 0.268 | 0.1974 | Yes |
| 19 | Ybx2 | 4067 | 0.250 | 0.1974 | Yes |
| 20 | Gfi1 | 4127 | 0.249 | 0.2085 | Yes |
| 21 | Nos1 | 4153 | 0.248 | 0.2213 | Yes |
| 22 | Sirt1 | 4163 | 0.247 | 0.2348 | Yes |
| 23 | Tcp11 | 4229 | 0.243 | 0.2453 | Yes |
| 24 | Nek2 | 4549 | 0.228 | 0.2423 | Yes |
| 25 | Ddx4 | 4906 | 0.210 | 0.2363 | Yes |
| 26 | Rfc4 | 4986 | 0.206 | 0.2439 | Yes |
| 27 | Mast2 | 5150 | 0.198 | 0.2469 | Yes |
| 28 | Dbf4 | 5824 | 0.168 | 0.2227 | No |
| 29 | Clvs1 | 6054 | 0.158 | 0.2201 | No |
| 30 | Ccnb2 | 6446 | 0.140 | 0.2084 | No |
| 31 | Map7 | 6478 | 0.138 | 0.2147 | No |
| 32 | Csnk2a2 | 6848 | 0.122 | 0.2031 | No |
| 33 | Pebp1 | 7265 | 0.112 | 0.1885 | No |
| 34 | Grm8 | 7469 | 0.104 | 0.1843 | No |
| 35 | Camk4 | 7509 | 0.103 | 0.1881 | No |
| 36 | Scg5 | 7914 | 0.087 | 0.1727 | No |
| 37 | Ezh2 | 8370 | 0.069 | 0.1538 | No |
| 38 | Tnni3 | 8412 | 0.067 | 0.1556 | No |
| 39 | Phkg2 | 8435 | 0.066 | 0.1582 | No |
| 40 | Cnih2 | 8661 | 0.057 | 0.1501 | No |
| 41 | Ldhc | 8841 | 0.050 | 0.1440 | No |
| 42 | Hspa4l | 8982 | 0.045 | 0.1395 | No |
| 43 | Jam3 | 9421 | 0.029 | 0.1191 | No |
| 44 | Topbp1 | 9447 | 0.028 | 0.1195 | No |
| 45 | Mlf1 | 9591 | 0.023 | 0.1136 | No |
| 46 | Mtor | 9618 | 0.022 | 0.1136 | No |
| 47 | Braf | 9974 | 0.010 | 0.0963 | No |
| 48 | Zc2hc1c | 10153 | 0.003 | 0.0876 | No |
| 49 | Rad17 | 10225 | 0.001 | 0.0841 | No |
| 50 | Cdk1 | 10637 | -0.006 | 0.0638 | No |
| 51 | Pgs1 | 10701 | -0.009 | 0.0611 | No |
| 52 | Bub1 | 11102 | -0.022 | 0.0423 | No |
| 53 | Nefh | 11229 | -0.027 | 0.0375 | No |
| 54 | Pias2 | 11802 | -0.047 | 0.0114 | No |
| 55 | Mllt10 | 11965 | -0.052 | 0.0063 | No |
| 56 | Kif2c | 12149 | -0.058 | 0.0004 | No |
| 57 | Pacrg | 12189 | -0.059 | 0.0018 | No |
| 58 | Agfg1 | 12373 | -0.065 | -0.0037 | No |
| 59 | Vdac3 | 12382 | -0.066 | -0.0004 | No |
| 60 | Parp2 | 12732 | -0.079 | -0.0134 | No |
| 61 | Arl4a | 12862 | -0.083 | -0.0152 | No |
| 62 | Cct6b | 12928 | -0.086 | -0.0136 | No |
| 63 | Ip6k1 | 13188 | -0.095 | -0.0212 | No |
| 64 | Lpin1 | 13200 | -0.095 | -0.0164 | No |
| 65 | Il12rb2 | 13272 | -0.098 | -0.0144 | No |
| 66 | Gstm5 | 13830 | -0.118 | -0.0357 | No |
| 67 | Tle4 | 13875 | -0.120 | -0.0311 | No |
| 68 | Clpb | 13880 | -0.120 | -0.0245 | No |
| 69 | Zc3h14 | 13906 | -0.121 | -0.0189 | No |
| 70 | Zpbp | 14006 | -0.125 | -0.0168 | No |
| 71 | Chfr | 14157 | -0.132 | -0.0169 | No |
| 72 | Strbp | 14358 | -0.140 | -0.0190 | No |
| 73 | Nf2 | 14429 | -0.142 | -0.0145 | No |
| 74 | Aurka | 14452 | -0.143 | -0.0075 | No |
| 75 | Ncaph | 14496 | -0.145 | -0.0014 | No |
| 76 | Stam2 | 14673 | -0.152 | -0.0017 | No |
| 77 | Spata6 | 14881 | -0.161 | -0.0029 | No |
| 78 | Nphp1 | 14953 | -0.165 | 0.0029 | No |
| 79 | Acrbp | 15216 | -0.175 | -0.0004 | No |
| 80 | Taldo1 | 15330 | -0.181 | 0.0042 | No |
| 81 | Crisp2 | 15495 | -0.184 | 0.0064 | No |
| 82 | Ide | 15600 | -0.189 | 0.0119 | No |
| 83 | Hspa1l | 15774 | -0.197 | 0.0144 | No |
| 84 | Ttk | 16040 | -0.211 | 0.0130 | No |
| 85 | Dmc1 | 16043 | -0.211 | 0.0249 | No |
| 86 | Psmg1 | 16572 | -0.237 | 0.0118 | No |
| 87 | Phf7 | 16823 | -0.251 | 0.0134 | No |
| 88 | Tulp2 | 16873 | -0.254 | 0.0253 | No |
| 89 | Art3 | 17671 | -0.302 | 0.0025 | No |
| 90 | Slc12a2 | 17939 | -0.319 | 0.0071 | No |
| 91 | Tekt2 | 18491 | -0.363 | -0.0000 | No |
| 92 | Gmcl1 | 19461 | -0.499 | -0.0204 | No |
| 93 | Cftr | 19991 | -0.860 | 0.0018 | No |