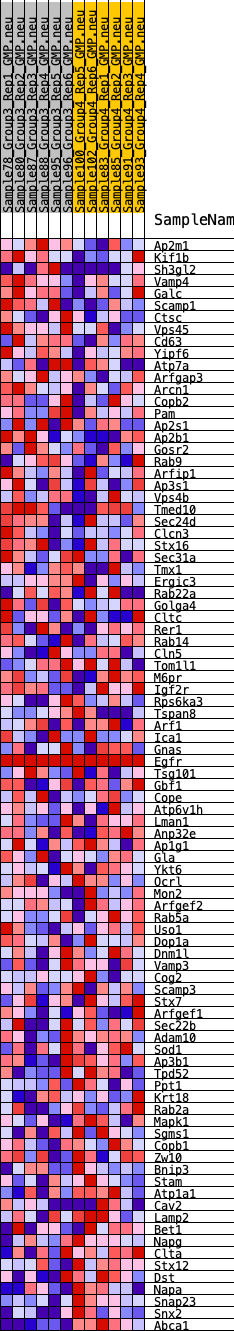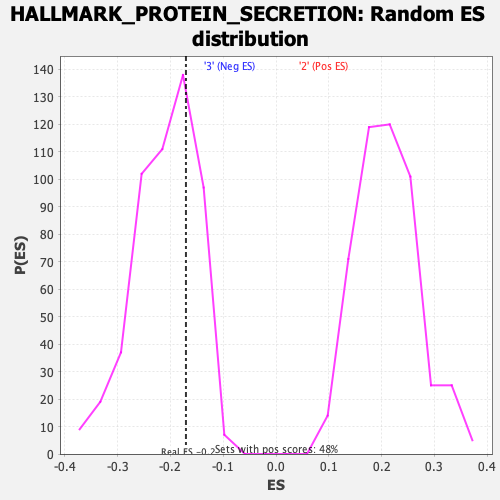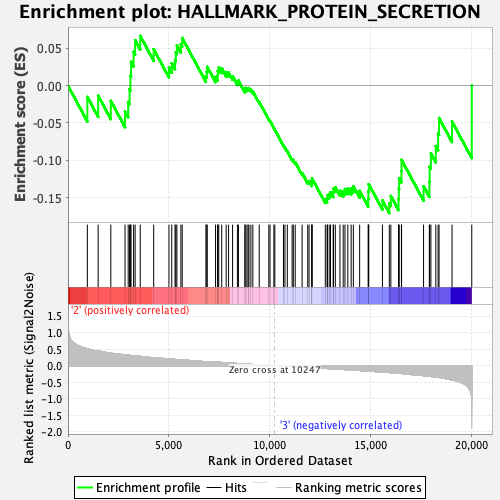
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group3_versus_Group4.GMP.neu_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | -0.1705757 |
| Normalized Enrichment Score (NES) | -0.8192389 |
| Nominal p-value | 0.68846154 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ap2m1 | 959 | 0.509 | -0.0151 | No |
| 2 | Kif1b | 1495 | 0.440 | -0.0135 | No |
| 3 | Sh3gl2 | 2123 | 0.379 | -0.0204 | No |
| 4 | Vamp4 | 2829 | 0.327 | -0.0346 | No |
| 5 | Galc | 2985 | 0.318 | -0.0217 | No |
| 6 | Scamp1 | 3053 | 0.313 | -0.0048 | No |
| 7 | Ctsc | 3095 | 0.311 | 0.0133 | No |
| 8 | Vps45 | 3126 | 0.310 | 0.0319 | No |
| 9 | Cd63 | 3253 | 0.304 | 0.0453 | No |
| 10 | Yipf6 | 3329 | 0.298 | 0.0608 | No |
| 11 | Atp7a | 3582 | 0.283 | 0.0665 | No |
| 12 | Arfgap3 | 4249 | 0.242 | 0.0487 | No |
| 13 | Arcn1 | 5006 | 0.204 | 0.0241 | No |
| 14 | Copb2 | 5146 | 0.198 | 0.0299 | No |
| 15 | Pam | 5309 | 0.190 | 0.0341 | No |
| 16 | Ap2s1 | 5347 | 0.188 | 0.0444 | No |
| 17 | Ap2b1 | 5400 | 0.186 | 0.0539 | No |
| 18 | Gosr2 | 5598 | 0.179 | 0.0556 | No |
| 19 | Rab9 | 5664 | 0.176 | 0.0637 | No |
| 20 | Arfip1 | 6840 | 0.122 | 0.0127 | No |
| 21 | Ap3s1 | 6884 | 0.121 | 0.0183 | No |
| 22 | Vps4b | 6902 | 0.120 | 0.0253 | No |
| 23 | Tmed10 | 7317 | 0.109 | 0.0116 | No |
| 24 | Sec24d | 7419 | 0.106 | 0.0134 | No |
| 25 | Clcn3 | 7429 | 0.106 | 0.0198 | No |
| 26 | Stx16 | 7476 | 0.104 | 0.0242 | No |
| 27 | Sec31a | 7624 | 0.099 | 0.0232 | No |
| 28 | Tmx1 | 7845 | 0.089 | 0.0180 | No |
| 29 | Ergic3 | 7964 | 0.085 | 0.0175 | No |
| 30 | Rab22a | 8162 | 0.077 | 0.0126 | No |
| 31 | Golga4 | 8402 | 0.068 | 0.0050 | No |
| 32 | Cltc | 8452 | 0.065 | 0.0068 | No |
| 33 | Rer1 | 8771 | 0.052 | -0.0058 | No |
| 34 | Rab14 | 8786 | 0.052 | -0.0031 | No |
| 35 | Cln5 | 8871 | 0.049 | -0.0042 | No |
| 36 | Tom1l1 | 8932 | 0.047 | -0.0041 | No |
| 37 | M6pr | 8974 | 0.045 | -0.0033 | No |
| 38 | Igf2r | 9071 | 0.041 | -0.0054 | No |
| 39 | Rps6ka3 | 9167 | 0.037 | -0.0078 | No |
| 40 | Tspan8 | 9484 | 0.027 | -0.0219 | No |
| 41 | Arf1 | 9959 | 0.011 | -0.0450 | No |
| 42 | Ica1 | 10018 | 0.009 | -0.0473 | No |
| 43 | Gnas | 10211 | 0.001 | -0.0569 | No |
| 44 | Egfr | 10256 | 0.000 | -0.0591 | No |
| 45 | Tsg101 | 10686 | -0.008 | -0.0801 | No |
| 46 | Gbf1 | 10758 | -0.011 | -0.0830 | No |
| 47 | Cope | 10876 | -0.015 | -0.0879 | No |
| 48 | Atp6v1h | 11115 | -0.023 | -0.0983 | No |
| 49 | Lman1 | 11173 | -0.025 | -0.0996 | No |
| 50 | Anp32e | 11268 | -0.028 | -0.1025 | No |
| 51 | Ap1g1 | 11610 | -0.040 | -0.1170 | No |
| 52 | Gla | 11889 | -0.050 | -0.1278 | No |
| 53 | Ykt6 | 11954 | -0.052 | -0.1276 | No |
| 54 | Ocrl | 12091 | -0.055 | -0.1308 | No |
| 55 | Mon2 | 12098 | -0.056 | -0.1275 | No |
| 56 | Arfgef2 | 12100 | -0.056 | -0.1240 | No |
| 57 | Rab5a | 12761 | -0.080 | -0.1519 | No |
| 58 | Uso1 | 12849 | -0.083 | -0.1509 | No |
| 59 | Dop1a | 12869 | -0.083 | -0.1465 | No |
| 60 | Dnm1l | 12954 | -0.087 | -0.1451 | No |
| 61 | Vamp3 | 13018 | -0.089 | -0.1425 | No |
| 62 | Cog2 | 13152 | -0.094 | -0.1431 | No |
| 63 | Scamp3 | 13165 | -0.094 | -0.1376 | No |
| 64 | Stx7 | 13256 | -0.097 | -0.1358 | No |
| 65 | Arfgef1 | 13487 | -0.105 | -0.1405 | No |
| 66 | Sec22b | 13647 | -0.111 | -0.1413 | No |
| 67 | Adam10 | 13735 | -0.115 | -0.1382 | No |
| 68 | Sod1 | 13877 | -0.120 | -0.1375 | No |
| 69 | Ap3b1 | 14042 | -0.127 | -0.1376 | No |
| 70 | Tpd52 | 14151 | -0.132 | -0.1345 | No |
| 71 | Ppt1 | 14464 | -0.144 | -0.1408 | No |
| 72 | Krt18 | 14888 | -0.162 | -0.1515 | No |
| 73 | Rab2a | 14898 | -0.162 | -0.1415 | No |
| 74 | Mapk1 | 14916 | -0.163 | -0.1318 | No |
| 75 | Sgms1 | 15594 | -0.188 | -0.1536 | No |
| 76 | Copb1 | 15934 | -0.205 | -0.1573 | Yes |
| 77 | Zw10 | 16011 | -0.210 | -0.1475 | Yes |
| 78 | Bnip3 | 16392 | -0.227 | -0.1519 | Yes |
| 79 | Stam | 16404 | -0.228 | -0.1377 | Yes |
| 80 | Atp1a1 | 16422 | -0.229 | -0.1238 | Yes |
| 81 | Cav2 | 16535 | -0.234 | -0.1142 | Yes |
| 82 | Lamp2 | 16543 | -0.235 | -0.0993 | Yes |
| 83 | Bet1 | 17633 | -0.299 | -0.1346 | Yes |
| 84 | Napg | 17920 | -0.318 | -0.1284 | Yes |
| 85 | Clta | 17936 | -0.319 | -0.1085 | Yes |
| 86 | Stx12 | 17996 | -0.322 | -0.0905 | Yes |
| 87 | Dst | 18241 | -0.339 | -0.0808 | Yes |
| 88 | Napa | 18355 | -0.349 | -0.0639 | Yes |
| 89 | Snap23 | 18401 | -0.353 | -0.0433 | Yes |
| 90 | Snx2 | 19045 | -0.426 | -0.0480 | Yes |
| 91 | Abca1 | 20024 | -1.500 | 0.0001 | Yes |