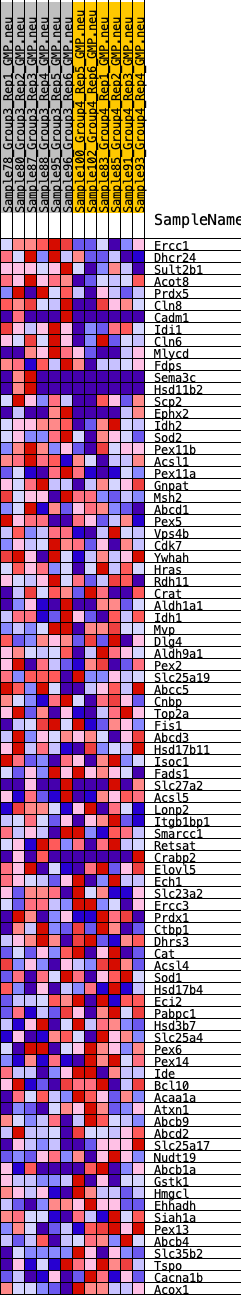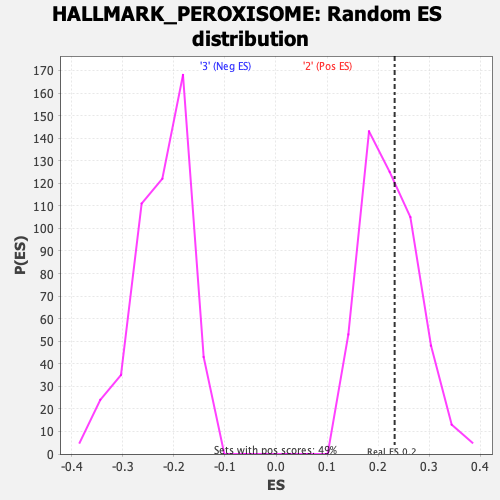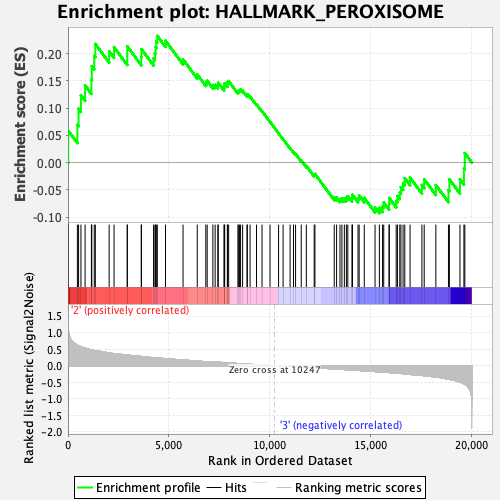
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group3_versus_Group4.GMP.neu_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | 0.23235266 |
| Normalized Enrichment Score (NES) | 1.0406933 |
| Nominal p-value | 0.38617885 |
| FDR q-value | 0.6803336 |
| FWER p-Value | 0.974 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ercc1 | 14 | 1.097 | 0.0583 | Yes |
| 2 | Dhcr24 | 465 | 0.621 | 0.0692 | Yes |
| 3 | Sult2b1 | 524 | 0.601 | 0.0986 | Yes |
| 4 | Acot8 | 642 | 0.569 | 0.1233 | Yes |
| 5 | Prdx5 | 847 | 0.530 | 0.1416 | Yes |
| 6 | Cln8 | 1160 | 0.484 | 0.1520 | Yes |
| 7 | Cadm1 | 1181 | 0.481 | 0.1769 | Yes |
| 8 | Idi1 | 1302 | 0.461 | 0.1957 | Yes |
| 9 | Cln6 | 1355 | 0.455 | 0.2175 | Yes |
| 10 | Mlycd | 2041 | 0.389 | 0.2041 | Yes |
| 11 | Fdps | 2284 | 0.367 | 0.2117 | Yes |
| 12 | Sema3c | 2939 | 0.319 | 0.1960 | Yes |
| 13 | Hsd11b2 | 2940 | 0.319 | 0.2132 | Yes |
| 14 | Scp2 | 3632 | 0.280 | 0.1936 | Yes |
| 15 | Ephx2 | 3643 | 0.279 | 0.2080 | Yes |
| 16 | Idh2 | 4243 | 0.242 | 0.1910 | Yes |
| 17 | Sod2 | 4310 | 0.238 | 0.2005 | Yes |
| 18 | Pex11b | 4344 | 0.236 | 0.2116 | Yes |
| 19 | Acsl1 | 4380 | 0.234 | 0.2224 | Yes |
| 20 | Pex11a | 4429 | 0.230 | 0.2324 | Yes |
| 21 | Gnpat | 4835 | 0.213 | 0.2235 | No |
| 22 | Msh2 | 5705 | 0.173 | 0.1893 | No |
| 23 | Abcd1 | 6410 | 0.141 | 0.1616 | No |
| 24 | Pex5 | 6834 | 0.123 | 0.1469 | No |
| 25 | Vps4b | 6902 | 0.120 | 0.1500 | No |
| 26 | Cdk7 | 7184 | 0.116 | 0.1422 | No |
| 27 | Ywhah | 7297 | 0.110 | 0.1425 | No |
| 28 | Hras | 7432 | 0.106 | 0.1414 | No |
| 29 | Rdh11 | 7449 | 0.105 | 0.1463 | No |
| 30 | Crat | 7739 | 0.094 | 0.1368 | No |
| 31 | Aldh1a1 | 7752 | 0.093 | 0.1413 | No |
| 32 | Idh1 | 7770 | 0.092 | 0.1454 | No |
| 33 | Mvp | 7903 | 0.087 | 0.1435 | No |
| 34 | Dlg4 | 7912 | 0.087 | 0.1477 | No |
| 35 | Aldh9a1 | 7967 | 0.085 | 0.1496 | No |
| 36 | Pex2 | 8426 | 0.067 | 0.1302 | No |
| 37 | Slc25a19 | 8467 | 0.065 | 0.1316 | No |
| 38 | Abcc5 | 8509 | 0.063 | 0.1330 | No |
| 39 | Cnbp | 8558 | 0.061 | 0.1339 | No |
| 40 | Top2a | 8660 | 0.057 | 0.1319 | No |
| 41 | Fis1 | 8879 | 0.049 | 0.1235 | No |
| 42 | Abcd3 | 8892 | 0.048 | 0.1255 | No |
| 43 | Hsd17b11 | 9029 | 0.043 | 0.1210 | No |
| 44 | Isoc1 | 9349 | 0.032 | 0.1067 | No |
| 45 | Fads1 | 9622 | 0.022 | 0.0942 | No |
| 46 | Slc27a2 | 10019 | 0.009 | 0.0748 | No |
| 47 | Acsl5 | 10443 | -0.000 | 0.0537 | No |
| 48 | Lonp2 | 10667 | -0.007 | 0.0429 | No |
| 49 | Itgb1bp1 | 11015 | -0.020 | 0.0265 | No |
| 50 | Smarcc1 | 11183 | -0.025 | 0.0195 | No |
| 51 | Retsat | 11286 | -0.029 | 0.0159 | No |
| 52 | Crabp2 | 11567 | -0.039 | 0.0039 | No |
| 53 | Elovl5 | 11819 | -0.047 | -0.0061 | No |
| 54 | Ech1 | 12209 | -0.060 | -0.0224 | No |
| 55 | Slc23a2 | 12246 | -0.061 | -0.0209 | No |
| 56 | Ercc3 | 13204 | -0.095 | -0.0638 | No |
| 57 | Prdx1 | 13321 | -0.099 | -0.0642 | No |
| 58 | Ctbp1 | 13488 | -0.105 | -0.0669 | No |
| 59 | Dhrs3 | 13584 | -0.109 | -0.0658 | No |
| 60 | Cat | 13705 | -0.114 | -0.0657 | No |
| 61 | Acsl4 | 13807 | -0.118 | -0.0644 | No |
| 62 | Sod1 | 13877 | -0.120 | -0.0614 | No |
| 63 | Hsd17b4 | 14082 | -0.128 | -0.0648 | No |
| 64 | Eci2 | 14109 | -0.130 | -0.0591 | No |
| 65 | Pabpc1 | 14384 | -0.141 | -0.0653 | No |
| 66 | Hsd3b7 | 14439 | -0.142 | -0.0603 | No |
| 67 | Slc25a4 | 14693 | -0.153 | -0.0648 | No |
| 68 | Pex6 | 15227 | -0.176 | -0.0821 | No |
| 69 | Pex14 | 15442 | -0.182 | -0.0830 | No |
| 70 | Ide | 15600 | -0.189 | -0.0807 | No |
| 71 | Bcl10 | 15658 | -0.191 | -0.0733 | No |
| 72 | Acaa1a | 15923 | -0.205 | -0.0755 | No |
| 73 | Atxn1 | 15931 | -0.205 | -0.0648 | No |
| 74 | Abcb9 | 16275 | -0.220 | -0.0702 | No |
| 75 | Abcd2 | 16339 | -0.223 | -0.0614 | No |
| 76 | Slc25a17 | 16454 | -0.230 | -0.0548 | No |
| 77 | Nudt19 | 16510 | -0.233 | -0.0450 | No |
| 78 | Abcb1a | 16618 | -0.239 | -0.0375 | No |
| 79 | Gstk1 | 16703 | -0.244 | -0.0285 | No |
| 80 | Hmgcl | 16964 | -0.261 | -0.0276 | No |
| 81 | Ehhadh | 17552 | -0.293 | -0.0412 | No |
| 82 | Siah1a | 17665 | -0.302 | -0.0306 | No |
| 83 | Pex13 | 18243 | -0.339 | -0.0413 | No |
| 84 | Abcb4 | 18868 | -0.408 | -0.0507 | No |
| 85 | Slc35b2 | 18913 | -0.412 | -0.0307 | No |
| 86 | Tspo | 19438 | -0.495 | -0.0304 | No |
| 87 | Cacna1b | 19635 | -0.548 | -0.0107 | No |
| 88 | Acox1 | 19679 | -0.563 | 0.0174 | No |