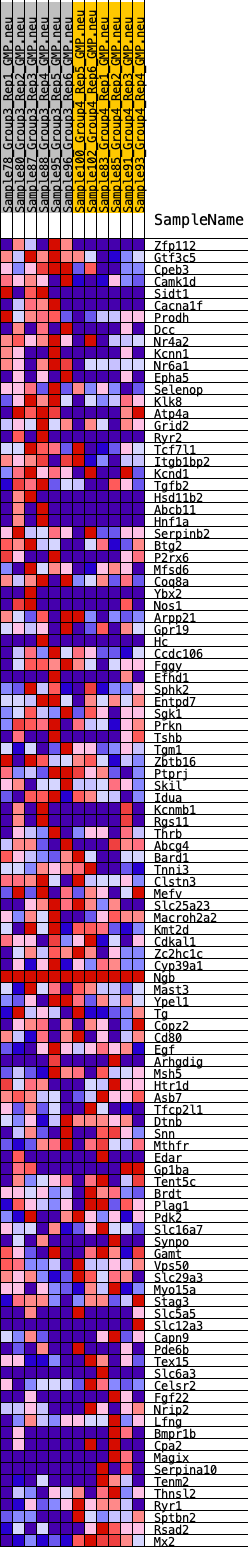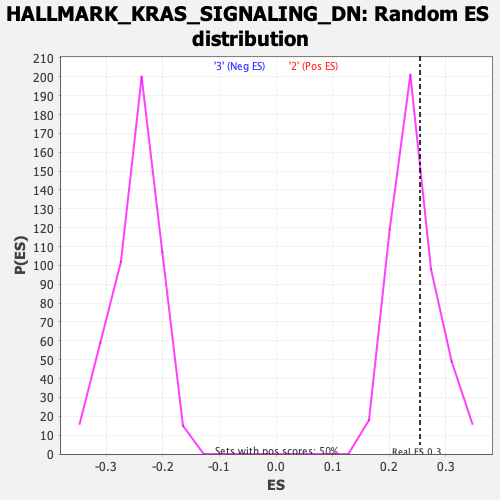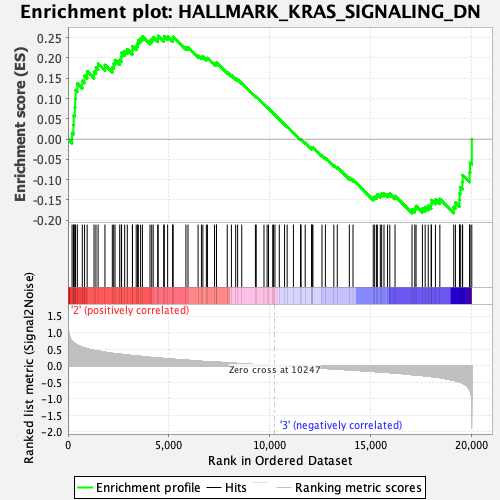
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group3_versus_Group4.GMP.neu_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.25453606 |
| Normalized Enrichment Score (NES) | 1.0468917 |
| Nominal p-value | 0.33333334 |
| FDR q-value | 0.70474946 |
| FWER p-Value | 0.974 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Zfp112 | 201 | 0.746 | 0.0151 | Yes |
| 2 | Gtf3c5 | 270 | 0.698 | 0.0352 | Yes |
| 3 | Cpeb3 | 282 | 0.694 | 0.0581 | Yes |
| 4 | Camk1d | 337 | 0.673 | 0.0781 | Yes |
| 5 | Sidt1 | 357 | 0.663 | 0.0995 | Yes |
| 6 | Cacna1f | 376 | 0.655 | 0.1207 | Yes |
| 7 | Prodh | 463 | 0.622 | 0.1374 | Yes |
| 8 | Dcc | 710 | 0.557 | 0.1439 | Yes |
| 9 | Nr4a2 | 821 | 0.537 | 0.1565 | Yes |
| 10 | Kcnn1 | 950 | 0.511 | 0.1673 | Yes |
| 11 | Nr6a1 | 1291 | 0.462 | 0.1658 | Yes |
| 12 | Epha5 | 1386 | 0.453 | 0.1764 | Yes |
| 13 | Selenop | 1493 | 0.440 | 0.1859 | Yes |
| 14 | Klk8 | 1832 | 0.409 | 0.1828 | Yes |
| 15 | Atp4a | 2198 | 0.372 | 0.1770 | Yes |
| 16 | Grid2 | 2267 | 0.369 | 0.1860 | Yes |
| 17 | Ryr2 | 2332 | 0.361 | 0.1950 | Yes |
| 18 | Tcf7l1 | 2564 | 0.346 | 0.1951 | Yes |
| 19 | Itgb1bp2 | 2646 | 0.341 | 0.2025 | Yes |
| 20 | Kcnd1 | 2658 | 0.340 | 0.2135 | Yes |
| 21 | Tgfb2 | 2810 | 0.329 | 0.2170 | Yes |
| 22 | Hsd11b2 | 2940 | 0.319 | 0.2213 | Yes |
| 23 | Abcb11 | 3195 | 0.305 | 0.2188 | Yes |
| 24 | Hnf1a | 3197 | 0.305 | 0.2290 | Yes |
| 25 | Serpinb2 | 3382 | 0.294 | 0.2297 | Yes |
| 26 | Btg2 | 3450 | 0.292 | 0.2362 | Yes |
| 27 | P2rx6 | 3488 | 0.289 | 0.2441 | Yes |
| 28 | Mfsd6 | 3600 | 0.282 | 0.2481 | Yes |
| 29 | Coq8a | 3690 | 0.275 | 0.2529 | Yes |
| 30 | Ybx2 | 4067 | 0.250 | 0.2425 | Yes |
| 31 | Nos1 | 4153 | 0.248 | 0.2466 | Yes |
| 32 | Arpp21 | 4226 | 0.244 | 0.2512 | Yes |
| 33 | Gpr19 | 4450 | 0.229 | 0.2477 | Yes |
| 34 | Hc | 4469 | 0.229 | 0.2545 | Yes |
| 35 | Ccdc106 | 4746 | 0.219 | 0.2481 | No |
| 36 | Fggy | 4776 | 0.216 | 0.2539 | No |
| 37 | Efhd1 | 4941 | 0.207 | 0.2527 | No |
| 38 | Sphk2 | 5186 | 0.196 | 0.2470 | No |
| 39 | Entpd7 | 5215 | 0.195 | 0.2522 | No |
| 40 | Sgk1 | 5848 | 0.167 | 0.2261 | No |
| 41 | Prkn | 5952 | 0.163 | 0.2264 | No |
| 42 | Tshb | 6450 | 0.140 | 0.2062 | No |
| 43 | Tgm1 | 6616 | 0.132 | 0.2024 | No |
| 44 | Zbtb16 | 6689 | 0.128 | 0.2031 | No |
| 45 | Ptprj | 6862 | 0.122 | 0.1985 | No |
| 46 | Skil | 6915 | 0.119 | 0.1999 | No |
| 47 | Idua | 7267 | 0.112 | 0.1861 | No |
| 48 | Kcnmb1 | 7363 | 0.108 | 0.1850 | No |
| 49 | Rgs11 | 7364 | 0.108 | 0.1886 | No |
| 50 | Thrb | 7897 | 0.088 | 0.1648 | No |
| 51 | Abcg4 | 8104 | 0.079 | 0.1572 | No |
| 52 | Bard1 | 8315 | 0.071 | 0.1490 | No |
| 53 | Tnni3 | 8412 | 0.067 | 0.1465 | No |
| 54 | Clstn3 | 8616 | 0.059 | 0.1382 | No |
| 55 | Mefv | 9294 | 0.034 | 0.1054 | No |
| 56 | Slc25a23 | 9334 | 0.032 | 0.1045 | No |
| 57 | Macroh2a2 | 9719 | 0.018 | 0.0859 | No |
| 58 | Kmt2d | 9874 | 0.013 | 0.0786 | No |
| 59 | Cdkal1 | 9943 | 0.011 | 0.0755 | No |
| 60 | Zc2hc1c | 10153 | 0.003 | 0.0651 | No |
| 61 | Cyp39a1 | 10195 | 0.002 | 0.0631 | No |
| 62 | Ngb | 10263 | 0.000 | 0.0598 | No |
| 63 | Mast3 | 10479 | -0.002 | 0.0490 | No |
| 64 | Ypel1 | 10739 | -0.010 | 0.0364 | No |
| 65 | Tg | 10868 | -0.015 | 0.0304 | No |
| 66 | Copz2 | 11179 | -0.025 | 0.0157 | No |
| 67 | Cd80 | 11530 | -0.037 | -0.0006 | No |
| 68 | Egf | 11559 | -0.038 | -0.0007 | No |
| 69 | Arhgdig | 11763 | -0.045 | -0.0094 | No |
| 70 | Msh5 | 12082 | -0.055 | -0.0235 | No |
| 71 | Htr1d | 12084 | -0.055 | -0.0217 | No |
| 72 | Asb7 | 12110 | -0.056 | -0.0210 | No |
| 73 | Tfcp2l1 | 12146 | -0.058 | -0.0209 | No |
| 74 | Dtnb | 12596 | -0.074 | -0.0409 | No |
| 75 | Snn | 12764 | -0.080 | -0.0466 | No |
| 76 | Mthfr | 13183 | -0.095 | -0.0644 | No |
| 77 | Edar | 13354 | -0.100 | -0.0696 | No |
| 78 | Gp1ba | 13952 | -0.123 | -0.0954 | No |
| 79 | Tent5c | 14134 | -0.131 | -0.1000 | No |
| 80 | Brdt | 15138 | -0.171 | -0.1446 | No |
| 81 | Plag1 | 15209 | -0.175 | -0.1423 | No |
| 82 | Pdk2 | 15304 | -0.180 | -0.1409 | No |
| 83 | Slc16a7 | 15341 | -0.182 | -0.1366 | No |
| 84 | Synpo | 15488 | -0.184 | -0.1377 | No |
| 85 | Gamt | 15555 | -0.186 | -0.1347 | No |
| 86 | Vps50 | 15671 | -0.192 | -0.1340 | No |
| 87 | Slc29a3 | 15847 | -0.201 | -0.1360 | No |
| 88 | Myo15a | 15958 | -0.206 | -0.1346 | No |
| 89 | Stag3 | 16218 | -0.217 | -0.1403 | No |
| 90 | Slc5a5 | 17061 | -0.267 | -0.1735 | No |
| 91 | Slc12a3 | 17199 | -0.272 | -0.1712 | No |
| 92 | Capn9 | 17260 | -0.274 | -0.1650 | No |
| 93 | Pde6b | 17576 | -0.295 | -0.1708 | No |
| 94 | Tex15 | 17713 | -0.305 | -0.1674 | No |
| 95 | Slc6a3 | 17865 | -0.316 | -0.1643 | No |
| 96 | Celsr2 | 18008 | -0.324 | -0.1605 | No |
| 97 | Fgf22 | 18028 | -0.326 | -0.1505 | No |
| 98 | Nrip2 | 18222 | -0.338 | -0.1487 | No |
| 99 | Lfng | 18441 | -0.357 | -0.1476 | No |
| 100 | Bmpr1b | 19130 | -0.438 | -0.1674 | No |
| 101 | Cpa2 | 19213 | -0.451 | -0.1563 | No |
| 102 | Magix | 19415 | -0.488 | -0.1499 | No |
| 103 | Serpina10 | 19420 | -0.488 | -0.1337 | No |
| 104 | Tenm2 | 19455 | -0.498 | -0.1186 | No |
| 105 | Thnsl2 | 19556 | -0.521 | -0.1060 | No |
| 106 | Ryr1 | 19571 | -0.526 | -0.0889 | No |
| 107 | Sptbn2 | 19915 | -0.707 | -0.0823 | No |
| 108 | Rsad2 | 19939 | -0.732 | -0.0587 | No |
| 109 | Mx2 | 20025 | -1.869 | 0.0001 | No |