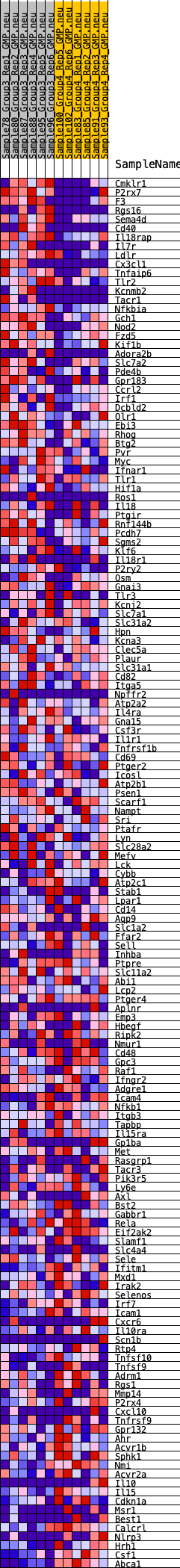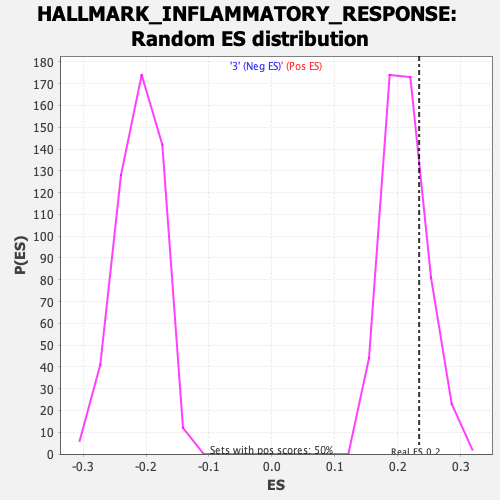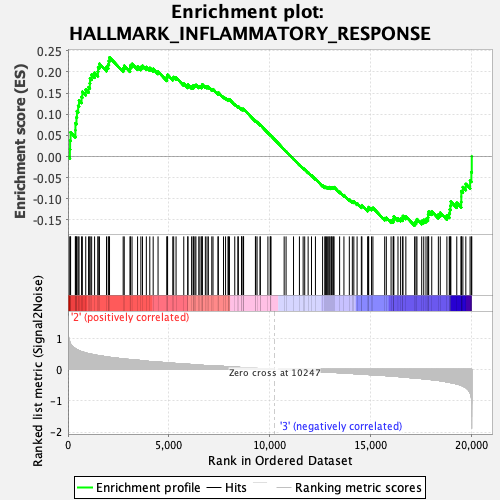
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group3_versus_Group4.GMP.neu_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_INFLAMMATORY_RESPONSE |
| Enrichment Score (ES) | 0.2342334 |
| Normalized Enrichment Score (NES) | 1.1074759 |
| Nominal p-value | 0.23742455 |
| FDR q-value | 0.6643088 |
| FWER p-Value | 0.962 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cmklr1 | 94 | 0.855 | 0.0170 | Yes |
| 2 | P2rx7 | 105 | 0.844 | 0.0379 | Yes |
| 3 | F3 | 129 | 0.804 | 0.0572 | Yes |
| 4 | Rgs16 | 366 | 0.660 | 0.0621 | Yes |
| 5 | Sema4d | 369 | 0.658 | 0.0787 | Yes |
| 6 | Cd40 | 420 | 0.638 | 0.0924 | Yes |
| 7 | Il18rap | 443 | 0.631 | 0.1073 | Yes |
| 8 | Il7r | 503 | 0.608 | 0.1198 | Yes |
| 9 | Ldlr | 547 | 0.595 | 0.1328 | Yes |
| 10 | Cx3cl1 | 673 | 0.564 | 0.1408 | Yes |
| 11 | Tnfaip6 | 711 | 0.556 | 0.1531 | Yes |
| 12 | Tlr2 | 880 | 0.527 | 0.1580 | Yes |
| 13 | Kcnmb2 | 1020 | 0.501 | 0.1637 | Yes |
| 14 | Tacr1 | 1082 | 0.491 | 0.1731 | Yes |
| 15 | Nfkbia | 1093 | 0.491 | 0.1851 | Yes |
| 16 | Gch1 | 1173 | 0.482 | 0.1934 | Yes |
| 17 | Nod2 | 1319 | 0.459 | 0.1977 | Yes |
| 18 | Fzd5 | 1483 | 0.441 | 0.2008 | Yes |
| 19 | Kif1b | 1495 | 0.440 | 0.2114 | Yes |
| 20 | Adora2b | 1560 | 0.434 | 0.2192 | Yes |
| 21 | Slc7a2 | 1909 | 0.400 | 0.2119 | Yes |
| 22 | Pde4b | 1995 | 0.394 | 0.2176 | Yes |
| 23 | Gpr183 | 2027 | 0.391 | 0.2260 | Yes |
| 24 | Ccrl2 | 2059 | 0.387 | 0.2342 | Yes |
| 25 | Irf1 | 2736 | 0.336 | 0.2088 | No |
| 26 | Dcbld2 | 2789 | 0.331 | 0.2145 | No |
| 27 | Olr1 | 3076 | 0.313 | 0.2081 | No |
| 28 | Ebi3 | 3094 | 0.311 | 0.2152 | No |
| 29 | Rhog | 3175 | 0.306 | 0.2189 | No |
| 30 | Btg2 | 3450 | 0.292 | 0.2125 | No |
| 31 | Pvr | 3613 | 0.281 | 0.2115 | No |
| 32 | Myc | 3693 | 0.275 | 0.2145 | No |
| 33 | Ifnar1 | 3885 | 0.261 | 0.2116 | No |
| 34 | Tlr1 | 4056 | 0.250 | 0.2094 | No |
| 35 | Hif1a | 4224 | 0.244 | 0.2072 | No |
| 36 | Ros1 | 4466 | 0.229 | 0.2008 | No |
| 37 | Il18 | 4907 | 0.210 | 0.1840 | No |
| 38 | Ptgir | 4912 | 0.209 | 0.1891 | No |
| 39 | Rnf144b | 4939 | 0.207 | 0.1931 | No |
| 40 | Pcdh7 | 5197 | 0.195 | 0.1851 | No |
| 41 | Sgms2 | 5242 | 0.194 | 0.1879 | No |
| 42 | Klf6 | 5359 | 0.188 | 0.1868 | No |
| 43 | Il18r1 | 5741 | 0.171 | 0.1720 | No |
| 44 | P2ry2 | 5939 | 0.163 | 0.1662 | No |
| 45 | Osm | 5948 | 0.163 | 0.1699 | No |
| 46 | Gnai3 | 6130 | 0.154 | 0.1648 | No |
| 47 | Tlr3 | 6211 | 0.150 | 0.1645 | No |
| 48 | Kcnj2 | 6215 | 0.150 | 0.1682 | No |
| 49 | Slc7a1 | 6292 | 0.146 | 0.1681 | No |
| 50 | Slc31a2 | 6343 | 0.143 | 0.1692 | No |
| 51 | Hpn | 6486 | 0.138 | 0.1656 | No |
| 52 | Kcna3 | 6544 | 0.134 | 0.1661 | No |
| 53 | Clec5a | 6632 | 0.130 | 0.1651 | No |
| 54 | Plaur | 6651 | 0.129 | 0.1674 | No |
| 55 | Slc31a1 | 6659 | 0.129 | 0.1704 | No |
| 56 | Cd82 | 6812 | 0.123 | 0.1658 | No |
| 57 | Itga5 | 6894 | 0.120 | 0.1648 | No |
| 58 | Npffr2 | 6961 | 0.119 | 0.1645 | No |
| 59 | Atp2a2 | 7131 | 0.118 | 0.1590 | No |
| 60 | Il4ra | 7191 | 0.115 | 0.1590 | No |
| 61 | Gna15 | 7439 | 0.105 | 0.1492 | No |
| 62 | Csf3r | 7453 | 0.105 | 0.1513 | No |
| 63 | Il1r1 | 7712 | 0.095 | 0.1407 | No |
| 64 | Tnfrsf1b | 7814 | 0.091 | 0.1379 | No |
| 65 | Cd69 | 7938 | 0.086 | 0.1339 | No |
| 66 | Ptger2 | 7954 | 0.085 | 0.1353 | No |
| 67 | Icosl | 8007 | 0.083 | 0.1348 | No |
| 68 | Atp2b1 | 8270 | 0.073 | 0.1235 | No |
| 69 | Psen1 | 8424 | 0.067 | 0.1175 | No |
| 70 | Scarf1 | 8447 | 0.066 | 0.1180 | No |
| 71 | Nampt | 8605 | 0.059 | 0.1116 | No |
| 72 | Sri | 8606 | 0.059 | 0.1131 | No |
| 73 | Ptafr | 8680 | 0.056 | 0.1109 | No |
| 74 | Lyn | 8697 | 0.056 | 0.1115 | No |
| 75 | Slc28a2 | 8708 | 0.055 | 0.1124 | No |
| 76 | Mefv | 9294 | 0.034 | 0.0838 | No |
| 77 | Lck | 9311 | 0.033 | 0.0839 | No |
| 78 | Cybb | 9376 | 0.031 | 0.0814 | No |
| 79 | Atp2c1 | 9526 | 0.026 | 0.0746 | No |
| 80 | Stab1 | 9536 | 0.025 | 0.0748 | No |
| 81 | Lpar1 | 9914 | 0.012 | 0.0561 | No |
| 82 | Cd14 | 10026 | 0.008 | 0.0507 | No |
| 83 | Aqp9 | 10046 | 0.007 | 0.0500 | No |
| 84 | Slc1a2 | 10066 | 0.007 | 0.0492 | No |
| 85 | Ffar2 | 10720 | -0.009 | 0.0165 | No |
| 86 | Sell | 10815 | -0.013 | 0.0121 | No |
| 87 | Inhba | 11184 | -0.025 | -0.0057 | No |
| 88 | Ptpre | 11477 | -0.035 | -0.0195 | No |
| 89 | Slc11a2 | 11661 | -0.042 | -0.0277 | No |
| 90 | Abi1 | 11739 | -0.045 | -0.0304 | No |
| 91 | Lcp2 | 11909 | -0.050 | -0.0377 | No |
| 92 | Ptger4 | 12076 | -0.055 | -0.0446 | No |
| 93 | Aplnr | 12271 | -0.062 | -0.0528 | No |
| 94 | Emp3 | 12626 | -0.075 | -0.0687 | No |
| 95 | Hbegf | 12734 | -0.079 | -0.0721 | No |
| 96 | Ripk2 | 12775 | -0.080 | -0.0721 | No |
| 97 | Nmur1 | 12824 | -0.081 | -0.0724 | No |
| 98 | Cd48 | 12878 | -0.084 | -0.0729 | No |
| 99 | Gpc3 | 12936 | -0.086 | -0.0736 | No |
| 100 | Raf1 | 12957 | -0.087 | -0.0724 | No |
| 101 | Ifngr2 | 13038 | -0.090 | -0.0742 | No |
| 102 | Adgre1 | 13063 | -0.090 | -0.0731 | No |
| 103 | Icam4 | 13104 | -0.091 | -0.0728 | No |
| 104 | Nfkb1 | 13161 | -0.094 | -0.0732 | No |
| 105 | Itgb3 | 13195 | -0.095 | -0.0725 | No |
| 106 | Tapbp | 13470 | -0.105 | -0.0836 | No |
| 107 | Il15ra | 13684 | -0.113 | -0.0915 | No |
| 108 | Gp1ba | 13952 | -0.123 | -0.1018 | No |
| 109 | Met | 14101 | -0.130 | -0.1059 | No |
| 110 | Rasgrp1 | 14175 | -0.132 | -0.1062 | No |
| 111 | Tacr3 | 14337 | -0.139 | -0.1108 | No |
| 112 | Pik3r5 | 14548 | -0.147 | -0.1176 | No |
| 113 | Ly6e | 14575 | -0.148 | -0.1152 | No |
| 114 | Axl | 14859 | -0.161 | -0.1253 | No |
| 115 | Bst2 | 14899 | -0.162 | -0.1232 | No |
| 116 | Gabbr1 | 14912 | -0.163 | -0.1196 | No |
| 117 | Rela | 15053 | -0.169 | -0.1224 | No |
| 118 | Eif2ak2 | 15121 | -0.171 | -0.1214 | No |
| 119 | Slamf1 | 15705 | -0.194 | -0.1458 | No |
| 120 | Slc4a4 | 15786 | -0.198 | -0.1448 | No |
| 121 | Sele | 16022 | -0.210 | -0.1513 | No |
| 122 | Ifitm1 | 16123 | -0.212 | -0.1510 | No |
| 123 | Mxd1 | 16146 | -0.213 | -0.1467 | No |
| 124 | Irak2 | 16158 | -0.214 | -0.1418 | No |
| 125 | Selenos | 16365 | -0.225 | -0.1464 | No |
| 126 | Irf7 | 16495 | -0.232 | -0.1470 | No |
| 127 | Icam1 | 16597 | -0.238 | -0.1461 | No |
| 128 | Cxcr6 | 16610 | -0.239 | -0.1406 | No |
| 129 | Il10ra | 16755 | -0.247 | -0.1416 | No |
| 130 | Scn1b | 17189 | -0.272 | -0.1564 | No |
| 131 | Rtp4 | 17257 | -0.274 | -0.1528 | No |
| 132 | Tnfsf10 | 17305 | -0.277 | -0.1482 | No |
| 133 | Tnfsf9 | 17541 | -0.292 | -0.1525 | No |
| 134 | Adrm1 | 17642 | -0.300 | -0.1500 | No |
| 135 | Rgs1 | 17754 | -0.307 | -0.1477 | No |
| 136 | Mmp14 | 17849 | -0.314 | -0.1445 | No |
| 137 | P2rx4 | 17860 | -0.315 | -0.1370 | No |
| 138 | Cxcl10 | 17887 | -0.316 | -0.1303 | No |
| 139 | Tnfrsf9 | 18039 | -0.327 | -0.1296 | No |
| 140 | Gpr132 | 18367 | -0.350 | -0.1371 | No |
| 141 | Ahr | 18462 | -0.359 | -0.1327 | No |
| 142 | Acvr1b | 18790 | -0.397 | -0.1391 | No |
| 143 | Sphk1 | 18909 | -0.411 | -0.1346 | No |
| 144 | Nmi | 18934 | -0.415 | -0.1253 | No |
| 145 | Acvr2a | 18967 | -0.419 | -0.1162 | No |
| 146 | Il10 | 18987 | -0.420 | -0.1065 | No |
| 147 | Il15 | 19280 | -0.461 | -0.1095 | No |
| 148 | Cdkn1a | 19498 | -0.508 | -0.1075 | No |
| 149 | Msr1 | 19504 | -0.509 | -0.0948 | No |
| 150 | Best1 | 19505 | -0.510 | -0.0819 | No |
| 151 | Calcrl | 19583 | -0.529 | -0.0723 | No |
| 152 | Nlrp3 | 19729 | -0.578 | -0.0649 | No |
| 153 | Hrh1 | 19940 | -0.733 | -0.0569 | No |
| 154 | Csf1 | 20004 | -0.905 | -0.0370 | No |
| 155 | Abca1 | 20024 | -1.500 | 0.0001 | No |