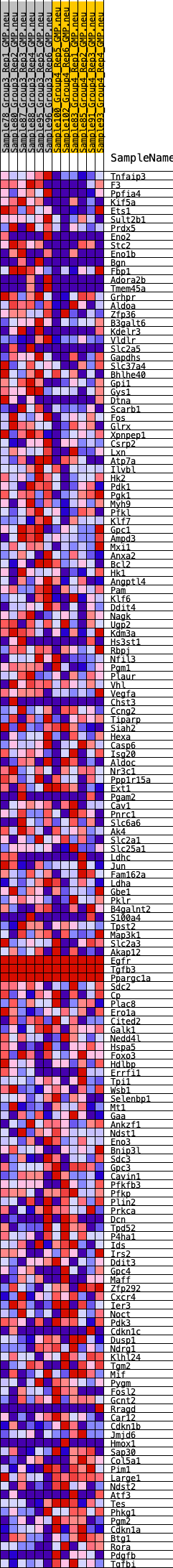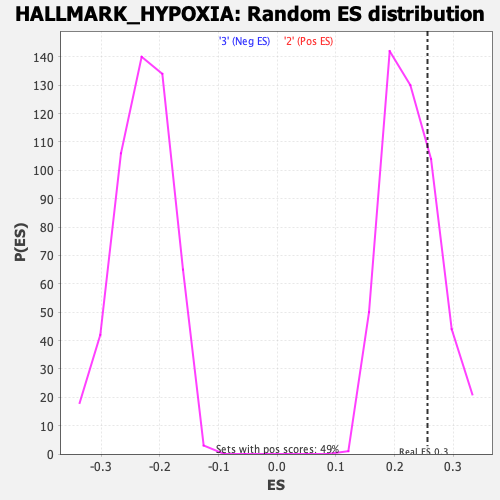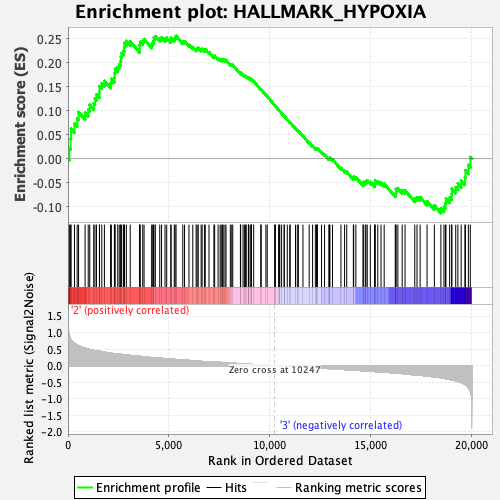
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group3_versus_Group4.GMP.neu_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_HYPOXIA |
| Enrichment Score (ES) | 0.25598404 |
| Normalized Enrichment Score (NES) | 1.1251258 |
| Nominal p-value | 0.26422763 |
| FDR q-value | 0.6592321 |
| FWER p-Value | 0.949 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tnfaip3 | 67 | 0.917 | 0.0223 | Yes |
| 2 | F3 | 129 | 0.804 | 0.0418 | Yes |
| 3 | Ppfia4 | 152 | 0.787 | 0.0627 | Yes |
| 4 | Kif5a | 321 | 0.680 | 0.0733 | Yes |
| 5 | Ets1 | 455 | 0.624 | 0.0841 | Yes |
| 6 | Sult2b1 | 524 | 0.601 | 0.0975 | Yes |
| 7 | Prdx5 | 847 | 0.530 | 0.0962 | Yes |
| 8 | Eno2 | 1003 | 0.502 | 0.1024 | Yes |
| 9 | Stc2 | 1078 | 0.492 | 0.1125 | Yes |
| 10 | Eno1b | 1276 | 0.465 | 0.1156 | Yes |
| 11 | Bgn | 1337 | 0.457 | 0.1254 | Yes |
| 12 | Fbp1 | 1412 | 0.450 | 0.1343 | Yes |
| 13 | Adora2b | 1560 | 0.434 | 0.1390 | Yes |
| 14 | Tmem45a | 1561 | 0.434 | 0.1512 | Yes |
| 15 | Grhpr | 1676 | 0.425 | 0.1574 | Yes |
| 16 | Aldoa | 1805 | 0.411 | 0.1625 | Yes |
| 17 | Zfp36 | 2112 | 0.380 | 0.1577 | Yes |
| 18 | B3galt6 | 2150 | 0.376 | 0.1664 | Yes |
| 19 | Kdelr3 | 2301 | 0.365 | 0.1691 | Yes |
| 20 | Vldlr | 2315 | 0.364 | 0.1786 | Yes |
| 21 | Slc2a5 | 2337 | 0.361 | 0.1877 | Yes |
| 22 | Gapdhs | 2467 | 0.355 | 0.1911 | Yes |
| 23 | Slc37a4 | 2541 | 0.348 | 0.1972 | Yes |
| 24 | Bhlhe40 | 2612 | 0.345 | 0.2033 | Yes |
| 25 | Gpi1 | 2626 | 0.343 | 0.2123 | Yes |
| 26 | Gys1 | 2662 | 0.340 | 0.2200 | Yes |
| 27 | Dtna | 2754 | 0.334 | 0.2248 | Yes |
| 28 | Scarb1 | 2792 | 0.331 | 0.2322 | Yes |
| 29 | Fos | 2798 | 0.330 | 0.2412 | Yes |
| 30 | Glrx | 2893 | 0.321 | 0.2455 | Yes |
| 31 | Xpnpep1 | 3086 | 0.312 | 0.2445 | Yes |
| 32 | Csrp2 | 3550 | 0.285 | 0.2292 | Yes |
| 33 | Lxn | 3553 | 0.285 | 0.2371 | Yes |
| 34 | Atp7a | 3582 | 0.283 | 0.2436 | Yes |
| 35 | Ilvbl | 3706 | 0.275 | 0.2451 | Yes |
| 36 | Hk2 | 3772 | 0.269 | 0.2494 | Yes |
| 37 | Pdk1 | 4148 | 0.248 | 0.2375 | Yes |
| 38 | Pgk1 | 4201 | 0.245 | 0.2417 | Yes |
| 39 | Myh9 | 4248 | 0.242 | 0.2462 | Yes |
| 40 | Pfkl | 4258 | 0.242 | 0.2525 | Yes |
| 41 | Klf7 | 4331 | 0.236 | 0.2555 | Yes |
| 42 | Gpc1 | 4550 | 0.228 | 0.2509 | Yes |
| 43 | Ampd3 | 4640 | 0.225 | 0.2527 | Yes |
| 44 | Mxi1 | 4812 | 0.215 | 0.2501 | Yes |
| 45 | Anxa2 | 4895 | 0.210 | 0.2519 | Yes |
| 46 | Bcl2 | 5101 | 0.200 | 0.2472 | Yes |
| 47 | Hk1 | 5111 | 0.200 | 0.2523 | Yes |
| 48 | Angptl4 | 5270 | 0.192 | 0.2498 | Yes |
| 49 | Pam | 5309 | 0.190 | 0.2532 | Yes |
| 50 | Klf6 | 5359 | 0.188 | 0.2560 | Yes |
| 51 | Ddit4 | 5693 | 0.174 | 0.2441 | No |
| 52 | Nagk | 5780 | 0.170 | 0.2445 | No |
| 53 | Ugp2 | 5999 | 0.161 | 0.2381 | No |
| 54 | Kdm3a | 6189 | 0.151 | 0.2328 | No |
| 55 | Hs3st1 | 6355 | 0.143 | 0.2285 | No |
| 56 | Rbpj | 6404 | 0.142 | 0.2300 | No |
| 57 | Nfil3 | 6459 | 0.139 | 0.2312 | No |
| 58 | Pgm1 | 6603 | 0.132 | 0.2277 | No |
| 59 | Plaur | 6651 | 0.129 | 0.2290 | No |
| 60 | Vhl | 6766 | 0.124 | 0.2267 | No |
| 61 | Vegfa | 6806 | 0.123 | 0.2282 | No |
| 62 | Chst3 | 6993 | 0.119 | 0.2222 | No |
| 63 | Ccng2 | 7235 | 0.113 | 0.2132 | No |
| 64 | Tiparp | 7273 | 0.111 | 0.2145 | No |
| 65 | Siah2 | 7445 | 0.105 | 0.2088 | No |
| 66 | Hexa | 7546 | 0.101 | 0.2066 | No |
| 67 | Casp6 | 7600 | 0.099 | 0.2067 | No |
| 68 | Isg20 | 7667 | 0.097 | 0.2061 | No |
| 69 | Aldoc | 7695 | 0.095 | 0.2075 | No |
| 70 | Nr3c1 | 7784 | 0.092 | 0.2056 | No |
| 71 | Ppp1r15a | 7835 | 0.090 | 0.2056 | No |
| 72 | Ext1 | 8051 | 0.081 | 0.1970 | No |
| 73 | Pgam2 | 8122 | 0.078 | 0.1957 | No |
| 74 | Cav1 | 8171 | 0.077 | 0.1954 | No |
| 75 | Pnrc1 | 8551 | 0.061 | 0.1781 | No |
| 76 | Slc6a6 | 8562 | 0.061 | 0.1793 | No |
| 77 | Ak4 | 8681 | 0.056 | 0.1749 | No |
| 78 | Slc2a1 | 8748 | 0.053 | 0.1731 | No |
| 79 | Slc25a1 | 8798 | 0.051 | 0.1721 | No |
| 80 | Ldhc | 8841 | 0.050 | 0.1714 | No |
| 81 | Jun | 8945 | 0.046 | 0.1675 | No |
| 82 | Fam162a | 8957 | 0.046 | 0.1682 | No |
| 83 | Ldha | 8969 | 0.045 | 0.1689 | No |
| 84 | Gbe1 | 9070 | 0.041 | 0.1650 | No |
| 85 | Pklr | 9079 | 0.041 | 0.1658 | No |
| 86 | B4galnt2 | 9097 | 0.040 | 0.1660 | No |
| 87 | S100a4 | 9208 | 0.036 | 0.1615 | No |
| 88 | Tpst2 | 9560 | 0.024 | 0.1445 | No |
| 89 | Map3k1 | 9581 | 0.023 | 0.1442 | No |
| 90 | Slc2a3 | 9815 | 0.015 | 0.1328 | No |
| 91 | Akap12 | 9879 | 0.013 | 0.1300 | No |
| 92 | Egfr | 10256 | 0.000 | 0.1111 | No |
| 93 | Tgfb3 | 10264 | 0.000 | 0.1108 | No |
| 94 | Ppargc1a | 10285 | 0.000 | 0.1098 | No |
| 95 | Sdc2 | 10444 | -0.001 | 0.1018 | No |
| 96 | Cp | 10481 | -0.002 | 0.1000 | No |
| 97 | Plac8 | 10503 | -0.002 | 0.0991 | No |
| 98 | Ero1a | 10594 | -0.005 | 0.0947 | No |
| 99 | Cited2 | 10719 | -0.009 | 0.0887 | No |
| 100 | Galk1 | 10727 | -0.009 | 0.0886 | No |
| 101 | Nedd4l | 10874 | -0.015 | 0.0817 | No |
| 102 | Hspa5 | 10997 | -0.019 | 0.0761 | No |
| 103 | Foxo3 | 11016 | -0.020 | 0.0757 | No |
| 104 | Hdlbp | 11282 | -0.029 | 0.0632 | No |
| 105 | Errfi1 | 11371 | -0.032 | 0.0596 | No |
| 106 | Tpi1 | 11424 | -0.034 | 0.0580 | No |
| 107 | Wsb1 | 11652 | -0.041 | 0.0477 | No |
| 108 | Selenbp1 | 11961 | -0.052 | 0.0337 | No |
| 109 | Mt1 | 12130 | -0.057 | 0.0268 | No |
| 110 | Gaa | 12266 | -0.062 | 0.0217 | No |
| 111 | Ankzf1 | 12325 | -0.064 | 0.0206 | No |
| 112 | Ndst1 | 12346 | -0.064 | 0.0214 | No |
| 113 | Eno3 | 12368 | -0.065 | 0.0222 | No |
| 114 | Bnip3l | 12573 | -0.073 | 0.0139 | No |
| 115 | Sdc3 | 12724 | -0.079 | 0.0086 | No |
| 116 | Gpc3 | 12936 | -0.086 | 0.0004 | No |
| 117 | Cavin1 | 12967 | -0.087 | 0.0013 | No |
| 118 | Pfkfb3 | 12989 | -0.088 | 0.0027 | No |
| 119 | Pfkp | 13118 | -0.092 | -0.0012 | No |
| 120 | Plin2 | 13537 | -0.107 | -0.0192 | No |
| 121 | Prkca | 13716 | -0.114 | -0.0250 | No |
| 122 | Dcn | 13823 | -0.118 | -0.0270 | No |
| 123 | Tpd52 | 14151 | -0.132 | -0.0398 | No |
| 124 | P4ha1 | 14168 | -0.132 | -0.0369 | No |
| 125 | Ids | 14275 | -0.137 | -0.0384 | No |
| 126 | Irs2 | 14636 | -0.150 | -0.0523 | No |
| 127 | Ddit3 | 14640 | -0.151 | -0.0482 | No |
| 128 | Gpc4 | 14740 | -0.155 | -0.0489 | No |
| 129 | Maff | 14775 | -0.156 | -0.0462 | No |
| 130 | Zfp292 | 14840 | -0.159 | -0.0450 | No |
| 131 | Cxcr4 | 14994 | -0.167 | -0.0480 | No |
| 132 | Ier3 | 15206 | -0.175 | -0.0537 | No |
| 133 | Noct | 15221 | -0.176 | -0.0495 | No |
| 134 | Pdk3 | 15231 | -0.176 | -0.0450 | No |
| 135 | Cdkn1c | 15364 | -0.182 | -0.0466 | No |
| 136 | Dusp1 | 15524 | -0.185 | -0.0494 | No |
| 137 | Ndrg1 | 15680 | -0.192 | -0.0518 | No |
| 138 | Klhl24 | 16229 | -0.218 | -0.0733 | No |
| 139 | Tgm2 | 16262 | -0.219 | -0.0688 | No |
| 140 | Mif | 16263 | -0.219 | -0.0626 | No |
| 141 | Pygm | 16358 | -0.225 | -0.0611 | No |
| 142 | Fosl2 | 16573 | -0.237 | -0.0652 | No |
| 143 | Gcnt2 | 16715 | -0.245 | -0.0654 | No |
| 144 | Rragd | 17197 | -0.272 | -0.0820 | No |
| 145 | Car12 | 17308 | -0.277 | -0.0798 | No |
| 146 | Cdkn1b | 17463 | -0.287 | -0.0795 | No |
| 147 | Jmjd6 | 17806 | -0.311 | -0.0880 | No |
| 148 | Hmox1 | 18169 | -0.335 | -0.0969 | No |
| 149 | Sap30 | 18493 | -0.363 | -0.1029 | No |
| 150 | Col5a1 | 18645 | -0.377 | -0.1000 | No |
| 151 | Pim1 | 18716 | -0.388 | -0.0926 | No |
| 152 | Large1 | 18743 | -0.392 | -0.0830 | No |
| 153 | Ndst2 | 18926 | -0.414 | -0.0805 | No |
| 154 | Atf3 | 19027 | -0.424 | -0.0737 | No |
| 155 | Tes | 19035 | -0.425 | -0.0621 | No |
| 156 | Phkg1 | 19228 | -0.455 | -0.0591 | No |
| 157 | Pgm2 | 19335 | -0.470 | -0.0512 | No |
| 158 | Cdkn1a | 19498 | -0.508 | -0.0451 | No |
| 159 | Btg1 | 19684 | -0.564 | -0.0387 | No |
| 160 | Rora | 19708 | -0.573 | -0.0238 | No |
| 161 | Pdgfb | 19856 | -0.654 | -0.0128 | No |
| 162 | Tgfbi | 19952 | -0.761 | 0.0037 | No |