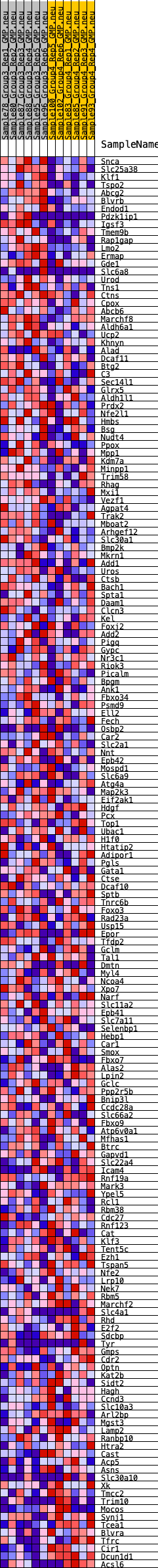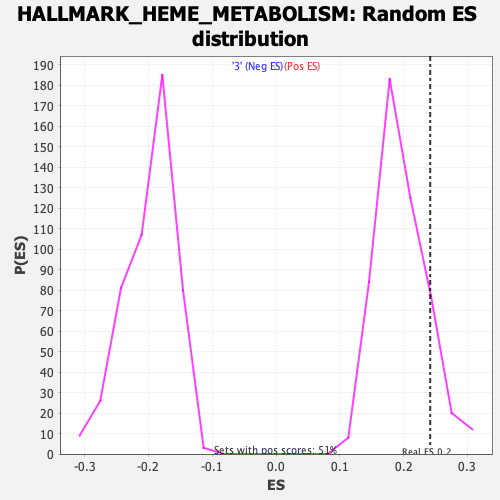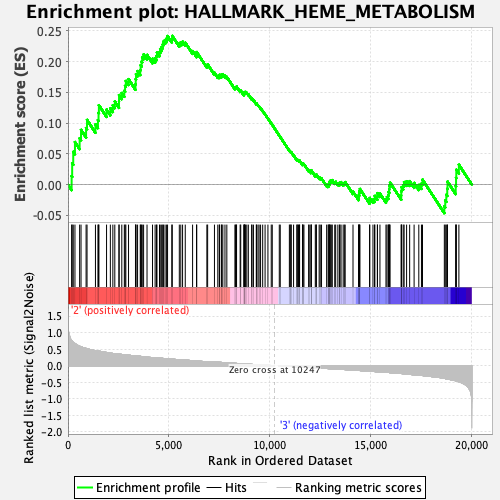
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group3_versus_Group4.GMP.neu_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_HEME_METABOLISM |
| Enrichment Score (ES) | 0.24181621 |
| Normalized Enrichment Score (NES) | 1.2332835 |
| Nominal p-value | 0.12770137 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.87 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Snca | 180 | 0.758 | 0.0137 | Yes |
| 2 | Slc25a38 | 207 | 0.742 | 0.0348 | Yes |
| 3 | Klf1 | 260 | 0.703 | 0.0533 | Yes |
| 4 | Tspo2 | 345 | 0.668 | 0.0692 | Yes |
| 5 | Abcg2 | 577 | 0.587 | 0.0753 | Yes |
| 6 | Blvrb | 646 | 0.569 | 0.0890 | Yes |
| 7 | Endod1 | 897 | 0.522 | 0.0921 | Yes |
| 8 | Pdzk1ip1 | 944 | 0.512 | 0.1052 | Yes |
| 9 | Igsf3 | 1361 | 0.454 | 0.0979 | Yes |
| 10 | Tmem9b | 1485 | 0.441 | 0.1050 | Yes |
| 11 | Rap1gap | 1515 | 0.438 | 0.1167 | Yes |
| 12 | Lmo2 | 1532 | 0.436 | 0.1290 | Yes |
| 13 | Ermap | 1912 | 0.400 | 0.1220 | Yes |
| 14 | Gde1 | 2100 | 0.381 | 0.1240 | Yes |
| 15 | Slc6a8 | 2224 | 0.370 | 0.1290 | Yes |
| 16 | Urod | 2321 | 0.363 | 0.1351 | Yes |
| 17 | Tns1 | 2529 | 0.349 | 0.1352 | Yes |
| 18 | Ctns | 2532 | 0.349 | 0.1456 | Yes |
| 19 | Cpox | 2668 | 0.339 | 0.1490 | Yes |
| 20 | Abcb6 | 2793 | 0.330 | 0.1527 | Yes |
| 21 | Marchf8 | 2831 | 0.327 | 0.1607 | Yes |
| 22 | Aldh6a1 | 2868 | 0.322 | 0.1686 | Yes |
| 23 | Ucp2 | 3002 | 0.317 | 0.1714 | Yes |
| 24 | Khnyn | 3346 | 0.297 | 0.1631 | Yes |
| 25 | Alad | 3358 | 0.296 | 0.1714 | Yes |
| 26 | Dcaf11 | 3374 | 0.295 | 0.1795 | Yes |
| 27 | Btg2 | 3450 | 0.292 | 0.1846 | Yes |
| 28 | C3 | 3583 | 0.283 | 0.1864 | Yes |
| 29 | Sec14l1 | 3602 | 0.282 | 0.1940 | Yes |
| 30 | Glrx5 | 3649 | 0.278 | 0.2001 | Yes |
| 31 | Aldh1l1 | 3680 | 0.276 | 0.2069 | Yes |
| 32 | Prdx2 | 3749 | 0.271 | 0.2116 | Yes |
| 33 | Nfe2l1 | 3919 | 0.259 | 0.2109 | Yes |
| 34 | Hmbs | 4192 | 0.245 | 0.2046 | Yes |
| 35 | Bsg | 4320 | 0.237 | 0.2054 | Yes |
| 36 | Nudt4 | 4387 | 0.233 | 0.2090 | Yes |
| 37 | Ppox | 4412 | 0.232 | 0.2148 | Yes |
| 38 | Mpp1 | 4537 | 0.229 | 0.2155 | Yes |
| 39 | Kdm7a | 4573 | 0.227 | 0.2205 | Yes |
| 40 | Minpp1 | 4645 | 0.225 | 0.2237 | Yes |
| 41 | Trim58 | 4697 | 0.221 | 0.2278 | Yes |
| 42 | Rhag | 4732 | 0.219 | 0.2327 | Yes |
| 43 | Mxi1 | 4812 | 0.215 | 0.2352 | Yes |
| 44 | Vezf1 | 4890 | 0.211 | 0.2377 | Yes |
| 45 | Agpat4 | 4937 | 0.207 | 0.2416 | Yes |
| 46 | Trak2 | 5148 | 0.198 | 0.2370 | Yes |
| 47 | Mboat2 | 5170 | 0.196 | 0.2418 | Yes |
| 48 | Arhgef12 | 5520 | 0.183 | 0.2298 | No |
| 49 | Slc30a1 | 5594 | 0.179 | 0.2315 | No |
| 50 | Bmp2k | 5682 | 0.175 | 0.2324 | No |
| 51 | Mkrn1 | 5815 | 0.168 | 0.2308 | No |
| 52 | Add1 | 6179 | 0.152 | 0.2170 | No |
| 53 | Uros | 6377 | 0.143 | 0.2114 | No |
| 54 | Ctsb | 6382 | 0.142 | 0.2155 | No |
| 55 | Bach1 | 6887 | 0.121 | 0.1937 | No |
| 56 | Spta1 | 6925 | 0.119 | 0.1955 | No |
| 57 | Daam1 | 7263 | 0.112 | 0.1819 | No |
| 58 | Clcn3 | 7429 | 0.106 | 0.1767 | No |
| 59 | Kel | 7502 | 0.103 | 0.1762 | No |
| 60 | Foxj2 | 7518 | 0.102 | 0.1785 | No |
| 61 | Add2 | 7607 | 0.099 | 0.1771 | No |
| 62 | Pigq | 7626 | 0.099 | 0.1791 | No |
| 63 | Gypc | 7678 | 0.097 | 0.1795 | No |
| 64 | Nr3c1 | 7784 | 0.092 | 0.1769 | No |
| 65 | Riok3 | 7877 | 0.088 | 0.1750 | No |
| 66 | Picalm | 8280 | 0.073 | 0.1569 | No |
| 67 | Bpgm | 8303 | 0.072 | 0.1579 | No |
| 68 | Ank1 | 8346 | 0.070 | 0.1579 | No |
| 69 | Fbxo34 | 8359 | 0.069 | 0.1594 | No |
| 70 | Psmd9 | 8537 | 0.062 | 0.1524 | No |
| 71 | Ell2 | 8566 | 0.061 | 0.1528 | No |
| 72 | Fech | 8725 | 0.055 | 0.1465 | No |
| 73 | Osbp2 | 8731 | 0.054 | 0.1479 | No |
| 74 | Car2 | 8747 | 0.054 | 0.1487 | No |
| 75 | Slc2a1 | 8748 | 0.053 | 0.1503 | No |
| 76 | Nnt | 8776 | 0.052 | 0.1506 | No |
| 77 | Epb42 | 8808 | 0.051 | 0.1505 | No |
| 78 | Mospd1 | 8868 | 0.049 | 0.1490 | No |
| 79 | Slc6a9 | 8943 | 0.046 | 0.1467 | No |
| 80 | Atg4a | 9107 | 0.040 | 0.1397 | No |
| 81 | Map2k3 | 9125 | 0.039 | 0.1400 | No |
| 82 | Eif2ak1 | 9196 | 0.036 | 0.1375 | No |
| 83 | Hdgf | 9342 | 0.032 | 0.1312 | No |
| 84 | Pcx | 9343 | 0.032 | 0.1322 | No |
| 85 | Top1 | 9420 | 0.029 | 0.1292 | No |
| 86 | Ubac1 | 9477 | 0.028 | 0.1272 | No |
| 87 | H1f0 | 9550 | 0.025 | 0.1243 | No |
| 88 | Htatip2 | 9655 | 0.021 | 0.1197 | No |
| 89 | Adipor1 | 9774 | 0.016 | 0.1143 | No |
| 90 | Pgls | 9915 | 0.012 | 0.1076 | No |
| 91 | Gata1 | 10079 | 0.006 | 0.0995 | No |
| 92 | Ctse | 10130 | 0.004 | 0.0971 | No |
| 93 | Dcaf10 | 10478 | -0.002 | 0.0797 | No |
| 94 | Sptb | 10526 | -0.003 | 0.0774 | No |
| 95 | Tnrc6b | 10978 | -0.018 | 0.0553 | No |
| 96 | Foxo3 | 11016 | -0.020 | 0.0540 | No |
| 97 | Rad23a | 11055 | -0.021 | 0.0527 | No |
| 98 | Usp15 | 11069 | -0.021 | 0.0527 | No |
| 99 | Epor | 11187 | -0.025 | 0.0475 | No |
| 100 | Tfdp2 | 11352 | -0.031 | 0.0402 | No |
| 101 | Gclm | 11360 | -0.032 | 0.0408 | No |
| 102 | Tal1 | 11401 | -0.033 | 0.0398 | No |
| 103 | Dmtn | 11429 | -0.034 | 0.0395 | No |
| 104 | Myl4 | 11475 | -0.035 | 0.0383 | No |
| 105 | Ncoa4 | 11486 | -0.036 | 0.0388 | No |
| 106 | Xpo7 | 11629 | -0.040 | 0.0329 | No |
| 107 | Narf | 11631 | -0.041 | 0.0341 | No |
| 108 | Slc11a2 | 11661 | -0.042 | 0.0339 | No |
| 109 | Epb41 | 11700 | -0.043 | 0.0332 | No |
| 110 | Slc7a11 | 11934 | -0.051 | 0.0231 | No |
| 111 | Selenbp1 | 11961 | -0.052 | 0.0233 | No |
| 112 | Hebp1 | 12056 | -0.054 | 0.0202 | No |
| 113 | Car1 | 12058 | -0.054 | 0.0218 | No |
| 114 | Smox | 12062 | -0.055 | 0.0233 | No |
| 115 | Fbxo7 | 12264 | -0.062 | 0.0150 | No |
| 116 | Alas2 | 12310 | -0.063 | 0.0147 | No |
| 117 | Lpin2 | 12314 | -0.063 | 0.0164 | No |
| 118 | Gclc | 12455 | -0.068 | 0.0114 | No |
| 119 | Ppp2r5b | 12508 | -0.071 | 0.0109 | No |
| 120 | Bnip3l | 12573 | -0.073 | 0.0099 | No |
| 121 | Ccdc28a | 12827 | -0.082 | -0.0004 | No |
| 122 | Slc66a2 | 12901 | -0.085 | -0.0015 | No |
| 123 | Fbxo9 | 12934 | -0.086 | -0.0005 | No |
| 124 | Atp6v0a1 | 12944 | -0.086 | 0.0016 | No |
| 125 | Mfhas1 | 12980 | -0.087 | 0.0025 | No |
| 126 | Btrc | 12982 | -0.087 | 0.0051 | No |
| 127 | Gapvd1 | 13009 | -0.089 | 0.0064 | No |
| 128 | Slc22a4 | 13079 | -0.091 | 0.0057 | No |
| 129 | Icam4 | 13104 | -0.091 | 0.0072 | No |
| 130 | Rnf19a | 13228 | -0.096 | 0.0039 | No |
| 131 | Mark3 | 13275 | -0.098 | 0.0046 | No |
| 132 | Ypel5 | 13383 | -0.102 | 0.0022 | No |
| 133 | Rcl1 | 13460 | -0.104 | 0.0015 | No |
| 134 | Rbm38 | 13485 | -0.105 | 0.0035 | No |
| 135 | Cdc27 | 13541 | -0.107 | 0.0040 | No |
| 136 | Rnf123 | 13646 | -0.111 | 0.0021 | No |
| 137 | Cat | 13705 | -0.114 | 0.0026 | No |
| 138 | Klf3 | 13746 | -0.115 | 0.0040 | No |
| 139 | Tent5c | 14134 | -0.131 | -0.0115 | No |
| 140 | Ezh1 | 14411 | -0.142 | -0.0212 | No |
| 141 | Tspan5 | 14419 | -0.142 | -0.0172 | No |
| 142 | Nfe2 | 14441 | -0.142 | -0.0140 | No |
| 143 | Lrp10 | 14453 | -0.143 | -0.0103 | No |
| 144 | Nek7 | 14478 | -0.144 | -0.0071 | No |
| 145 | Rbm5 | 14955 | -0.165 | -0.0261 | No |
| 146 | Marchf2 | 14967 | -0.165 | -0.0217 | No |
| 147 | Slc4a1 | 15124 | -0.171 | -0.0244 | No |
| 148 | Rhd | 15205 | -0.174 | -0.0232 | No |
| 149 | E2f2 | 15215 | -0.175 | -0.0184 | No |
| 150 | Sdcbp | 15342 | -0.182 | -0.0193 | No |
| 151 | Tyr | 15347 | -0.182 | -0.0140 | No |
| 152 | Gmps | 15470 | -0.183 | -0.0146 | No |
| 153 | Cdr2 | 15769 | -0.197 | -0.0237 | No |
| 154 | Optn | 15819 | -0.200 | -0.0202 | No |
| 155 | Kat2b | 15885 | -0.203 | -0.0173 | No |
| 156 | Sidt2 | 15901 | -0.204 | -0.0120 | No |
| 157 | Hagh | 15933 | -0.205 | -0.0074 | No |
| 158 | Ccnd3 | 15939 | -0.206 | -0.0014 | No |
| 159 | Slc10a3 | 15973 | -0.207 | 0.0032 | No |
| 160 | Arl2bp | 16526 | -0.234 | -0.0176 | No |
| 161 | Mgst3 | 16529 | -0.234 | -0.0107 | No |
| 162 | Lamp2 | 16543 | -0.235 | -0.0043 | No |
| 163 | Ranbp10 | 16623 | -0.240 | -0.0010 | No |
| 164 | Htra2 | 16668 | -0.242 | 0.0041 | No |
| 165 | Cast | 16792 | -0.249 | 0.0054 | No |
| 166 | Acp5 | 16944 | -0.259 | 0.0056 | No |
| 167 | Asns | 17164 | -0.271 | 0.0027 | No |
| 168 | Slc30a10 | 17397 | -0.282 | -0.0005 | No |
| 169 | Xk | 17529 | -0.292 | 0.0017 | No |
| 170 | Tmcc2 | 17580 | -0.295 | 0.0080 | No |
| 171 | Trim10 | 18663 | -0.379 | -0.0350 | No |
| 172 | Mocos | 18709 | -0.386 | -0.0257 | No |
| 173 | Synj1 | 18761 | -0.394 | -0.0164 | No |
| 174 | Tcea1 | 18808 | -0.399 | -0.0067 | No |
| 175 | Blvra | 18821 | -0.401 | 0.0048 | No |
| 176 | Tfrc | 19227 | -0.454 | -0.0019 | No |
| 177 | Cir1 | 19245 | -0.456 | 0.0110 | No |
| 178 | Dcun1d1 | 19257 | -0.458 | 0.0242 | No |
| 179 | Acsl6 | 19385 | -0.481 | 0.0323 | No |