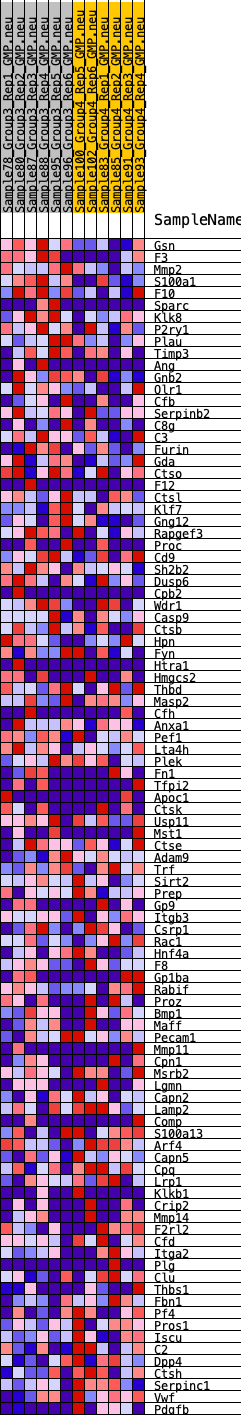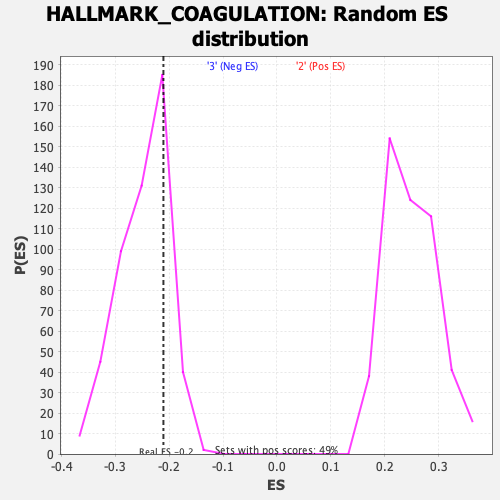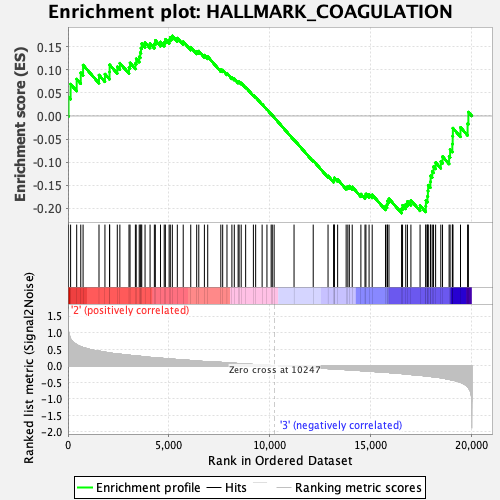
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group3_versus_Group4.GMP.neu_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | -0.2108809 |
| Normalized Enrichment Score (NES) | -0.851746 |
| Nominal p-value | 0.7749511 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gsn | 27 | 1.038 | 0.0413 | No |
| 2 | F3 | 129 | 0.804 | 0.0693 | No |
| 3 | Mmp2 | 431 | 0.635 | 0.0802 | No |
| 4 | S100a1 | 633 | 0.574 | 0.0937 | No |
| 5 | F10 | 748 | 0.549 | 0.1106 | No |
| 6 | Sparc | 1537 | 0.435 | 0.0889 | No |
| 7 | Klk8 | 1832 | 0.409 | 0.0910 | No |
| 8 | P2ry1 | 2060 | 0.387 | 0.0955 | No |
| 9 | Plau | 2066 | 0.386 | 0.1111 | No |
| 10 | Timp3 | 2444 | 0.356 | 0.1068 | No |
| 11 | Ang | 2572 | 0.346 | 0.1147 | No |
| 12 | Gnb2 | 3025 | 0.315 | 0.1049 | No |
| 13 | Olr1 | 3076 | 0.313 | 0.1153 | No |
| 14 | Cfb | 3342 | 0.297 | 0.1142 | No |
| 15 | Serpinb2 | 3382 | 0.294 | 0.1243 | No |
| 16 | C8g | 3532 | 0.287 | 0.1286 | No |
| 17 | C3 | 3583 | 0.283 | 0.1377 | No |
| 18 | Furin | 3618 | 0.280 | 0.1476 | No |
| 19 | Gda | 3657 | 0.277 | 0.1571 | No |
| 20 | Ctso | 3821 | 0.265 | 0.1598 | No |
| 21 | F12 | 4070 | 0.250 | 0.1576 | No |
| 22 | Ctsl | 4283 | 0.240 | 0.1568 | No |
| 23 | Klf7 | 4331 | 0.236 | 0.1642 | No |
| 24 | Gng12 | 4587 | 0.226 | 0.1607 | No |
| 25 | Rapgef3 | 4772 | 0.217 | 0.1604 | No |
| 26 | Proc | 4829 | 0.214 | 0.1664 | No |
| 27 | Cd9 | 5015 | 0.204 | 0.1654 | No |
| 28 | Sh2b2 | 5073 | 0.201 | 0.1709 | No |
| 29 | Dusp6 | 5174 | 0.196 | 0.1739 | No |
| 30 | Cpb2 | 5421 | 0.184 | 0.1691 | No |
| 31 | Wdr1 | 5714 | 0.173 | 0.1616 | No |
| 32 | Casp9 | 6087 | 0.156 | 0.1493 | No |
| 33 | Ctsb | 6382 | 0.142 | 0.1405 | No |
| 34 | Hpn | 6486 | 0.138 | 0.1409 | No |
| 35 | Fyn | 6764 | 0.125 | 0.1322 | No |
| 36 | Htra1 | 6929 | 0.119 | 0.1288 | No |
| 37 | Hmgcs2 | 7578 | 0.100 | 0.1004 | No |
| 38 | Thbd | 7671 | 0.097 | 0.0998 | No |
| 39 | Masp2 | 7884 | 0.088 | 0.0928 | No |
| 40 | Cfh | 8128 | 0.078 | 0.0838 | No |
| 41 | Anxa1 | 8247 | 0.074 | 0.0809 | No |
| 42 | Pef1 | 8437 | 0.066 | 0.0741 | No |
| 43 | Lta4h | 8485 | 0.064 | 0.0744 | No |
| 44 | Plek | 8589 | 0.060 | 0.0717 | No |
| 45 | Fn1 | 8810 | 0.051 | 0.0627 | No |
| 46 | Tfpi2 | 9194 | 0.036 | 0.0450 | No |
| 47 | Apoc1 | 9306 | 0.033 | 0.0408 | No |
| 48 | Ctsk | 9627 | 0.022 | 0.0256 | No |
| 49 | Usp11 | 9866 | 0.013 | 0.0142 | No |
| 50 | Mst1 | 10075 | 0.006 | 0.0041 | No |
| 51 | Ctse | 10130 | 0.004 | 0.0015 | No |
| 52 | Adam9 | 10223 | 0.001 | -0.0031 | No |
| 53 | Trf | 11210 | -0.026 | -0.0515 | No |
| 54 | Sirt2 | 12161 | -0.058 | -0.0967 | No |
| 55 | Prep | 12896 | -0.085 | -0.1301 | No |
| 56 | Gp9 | 13174 | -0.094 | -0.1401 | No |
| 57 | Itgb3 | 13195 | -0.095 | -0.1372 | No |
| 58 | Csrp1 | 13210 | -0.096 | -0.1340 | No |
| 59 | Rac1 | 13372 | -0.101 | -0.1379 | No |
| 60 | Hnf4a | 13794 | -0.117 | -0.1542 | No |
| 61 | F8 | 13878 | -0.120 | -0.1535 | No |
| 62 | Gp1ba | 13952 | -0.123 | -0.1521 | No |
| 63 | Rabif | 14095 | -0.129 | -0.1539 | No |
| 64 | Proz | 14525 | -0.146 | -0.1694 | No |
| 65 | Bmp1 | 14729 | -0.154 | -0.1733 | No |
| 66 | Maff | 14775 | -0.156 | -0.1691 | No |
| 67 | Pecam1 | 14931 | -0.164 | -0.1702 | No |
| 68 | Mmp11 | 15086 | -0.170 | -0.1709 | No |
| 69 | Cpn1 | 15745 | -0.195 | -0.1959 | No |
| 70 | Msrb2 | 15821 | -0.200 | -0.1915 | No |
| 71 | Lgmn | 15844 | -0.201 | -0.1843 | No |
| 72 | Capn2 | 15918 | -0.204 | -0.1796 | No |
| 73 | Lamp2 | 16543 | -0.235 | -0.2012 | Yes |
| 74 | Comp | 16591 | -0.238 | -0.1938 | Yes |
| 75 | S100a13 | 16750 | -0.247 | -0.1916 | Yes |
| 76 | Arf4 | 16834 | -0.251 | -0.1854 | Yes |
| 77 | Capn5 | 17006 | -0.263 | -0.1832 | Yes |
| 78 | Cpq | 17460 | -0.286 | -0.1941 | Yes |
| 79 | Lrp1 | 17743 | -0.306 | -0.1957 | Yes |
| 80 | Klkb1 | 17750 | -0.307 | -0.1834 | Yes |
| 81 | Crip2 | 17827 | -0.312 | -0.1744 | Yes |
| 82 | Mmp14 | 17849 | -0.314 | -0.1625 | Yes |
| 83 | F2rl2 | 17861 | -0.315 | -0.1501 | Yes |
| 84 | Cfd | 17972 | -0.320 | -0.1425 | Yes |
| 85 | Itga2 | 17984 | -0.321 | -0.1298 | Yes |
| 86 | Plg | 18063 | -0.328 | -0.1202 | Yes |
| 87 | Clu | 18128 | -0.333 | -0.1098 | Yes |
| 88 | Thbs1 | 18231 | -0.339 | -0.1010 | Yes |
| 89 | Fbn1 | 18488 | -0.363 | -0.0989 | Yes |
| 90 | Pf4 | 18574 | -0.370 | -0.0880 | Yes |
| 91 | Pros1 | 18900 | -0.409 | -0.0875 | Yes |
| 92 | Iscu | 18949 | -0.417 | -0.0728 | Yes |
| 93 | C2 | 19060 | -0.429 | -0.0606 | Yes |
| 94 | Dpp4 | 19077 | -0.432 | -0.0437 | Yes |
| 95 | Ctsh | 19087 | -0.434 | -0.0263 | Yes |
| 96 | Serpinc1 | 19465 | -0.500 | -0.0246 | Yes |
| 97 | Vwf | 19817 | -0.628 | -0.0165 | Yes |
| 98 | Pdgfb | 19856 | -0.654 | 0.0085 | Yes |