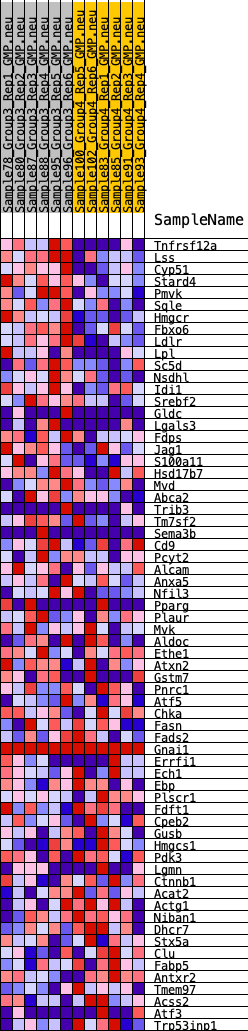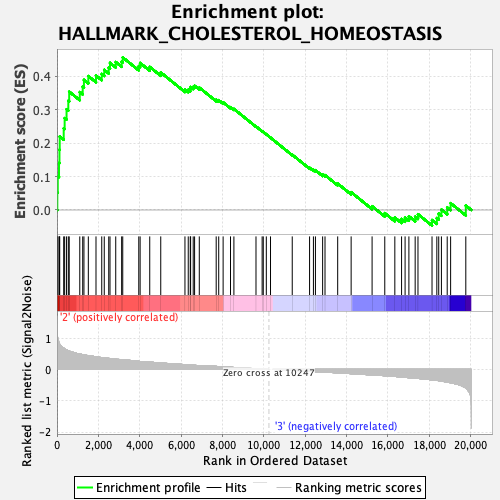
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group3_versus_Group4.GMP.neu_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | 0.4559038 |
| Normalized Enrichment Score (NES) | 1.6339138 |
| Nominal p-value | 0.0020283975 |
| FDR q-value | 0.41307592 |
| FWER p-Value | 0.203 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tnfrsf12a | 16 | 1.064 | 0.0523 | Yes |
| 2 | Lss | 37 | 1.000 | 0.1011 | Yes |
| 3 | Cyp51 | 98 | 0.852 | 0.1406 | Yes |
| 4 | Stard4 | 119 | 0.814 | 0.1802 | Yes |
| 5 | Pmvk | 131 | 0.802 | 0.2196 | Yes |
| 6 | Sqle | 324 | 0.680 | 0.2439 | Yes |
| 7 | Hmgcr | 370 | 0.658 | 0.2745 | Yes |
| 8 | Fbxo6 | 462 | 0.623 | 0.3010 | Yes |
| 9 | Ldlr | 547 | 0.595 | 0.3265 | Yes |
| 10 | Lpl | 590 | 0.585 | 0.3535 | Yes |
| 11 | Sc5d | 1101 | 0.488 | 0.3523 | Yes |
| 12 | Nsdhl | 1243 | 0.471 | 0.3688 | Yes |
| 13 | Idi1 | 1302 | 0.461 | 0.3889 | Yes |
| 14 | Srebf2 | 1514 | 0.438 | 0.4001 | Yes |
| 15 | Gldc | 1885 | 0.402 | 0.4017 | Yes |
| 16 | Lgals3 | 2163 | 0.374 | 0.4064 | Yes |
| 17 | Fdps | 2284 | 0.367 | 0.4187 | Yes |
| 18 | Jag1 | 2501 | 0.352 | 0.4254 | Yes |
| 19 | S100a11 | 2562 | 0.346 | 0.4397 | Yes |
| 20 | Hsd17b7 | 2839 | 0.325 | 0.4421 | Yes |
| 21 | Mvd | 3125 | 0.310 | 0.4433 | Yes |
| 22 | Abca2 | 3178 | 0.306 | 0.4559 | Yes |
| 23 | Trib3 | 3953 | 0.257 | 0.4299 | No |
| 24 | Tm7sf2 | 4018 | 0.252 | 0.4393 | No |
| 25 | Sema3b | 4483 | 0.229 | 0.4275 | No |
| 26 | Cd9 | 5015 | 0.204 | 0.4110 | No |
| 27 | Pcyt2 | 6185 | 0.151 | 0.3600 | No |
| 28 | Alcam | 6344 | 0.143 | 0.3593 | No |
| 29 | Anxa5 | 6441 | 0.140 | 0.3614 | No |
| 30 | Nfil3 | 6459 | 0.139 | 0.3675 | No |
| 31 | Pparg | 6584 | 0.133 | 0.3679 | No |
| 32 | Plaur | 6651 | 0.129 | 0.3711 | No |
| 33 | Mvk | 6878 | 0.121 | 0.3658 | No |
| 34 | Aldoc | 7695 | 0.095 | 0.3296 | No |
| 35 | Ethe1 | 7817 | 0.091 | 0.3281 | No |
| 36 | Atxn2 | 8030 | 0.082 | 0.3216 | No |
| 37 | Gstm7 | 8387 | 0.068 | 0.3072 | No |
| 38 | Pnrc1 | 8551 | 0.061 | 0.3021 | No |
| 39 | Atf5 | 9619 | 0.022 | 0.2497 | No |
| 40 | Chka | 9916 | 0.012 | 0.2355 | No |
| 41 | Fasn | 9971 | 0.010 | 0.2333 | No |
| 42 | Fads2 | 10118 | 0.005 | 0.2262 | No |
| 43 | Gnai1 | 10321 | 0.000 | 0.2161 | No |
| 44 | Errfi1 | 11371 | -0.032 | 0.1651 | No |
| 45 | Ech1 | 12209 | -0.060 | 0.1262 | No |
| 46 | Ebp | 12395 | -0.066 | 0.1202 | No |
| 47 | Plscr1 | 12492 | -0.070 | 0.1189 | No |
| 48 | Fdft1 | 12840 | -0.082 | 0.1056 | No |
| 49 | Cpeb2 | 12955 | -0.087 | 0.1042 | No |
| 50 | Gusb | 13568 | -0.108 | 0.0789 | No |
| 51 | Hmgcs1 | 14218 | -0.134 | 0.0531 | No |
| 52 | Pdk3 | 15231 | -0.176 | 0.0112 | No |
| 53 | Lgmn | 15844 | -0.201 | -0.0094 | No |
| 54 | Ctnnb1 | 16330 | -0.223 | -0.0226 | No |
| 55 | Acat2 | 16656 | -0.242 | -0.0268 | No |
| 56 | Actg1 | 16826 | -0.251 | -0.0228 | No |
| 57 | Niban1 | 17010 | -0.263 | -0.0188 | No |
| 58 | Dhcr7 | 17313 | -0.277 | -0.0201 | No |
| 59 | Stx5a | 17449 | -0.285 | -0.0126 | No |
| 60 | Clu | 18128 | -0.333 | -0.0300 | No |
| 61 | Fabp5 | 18360 | -0.349 | -0.0242 | No |
| 62 | Antxr2 | 18452 | -0.358 | -0.0109 | No |
| 63 | Tmem97 | 18582 | -0.371 | 0.0012 | No |
| 64 | Acss2 | 18862 | -0.406 | 0.0075 | No |
| 65 | Atf3 | 19027 | -0.424 | 0.0204 | No |
| 66 | Trp53inp1 | 19761 | -0.594 | 0.0133 | No |