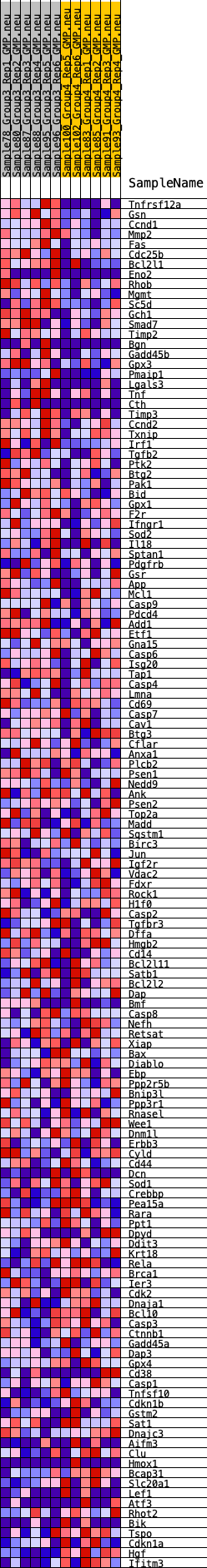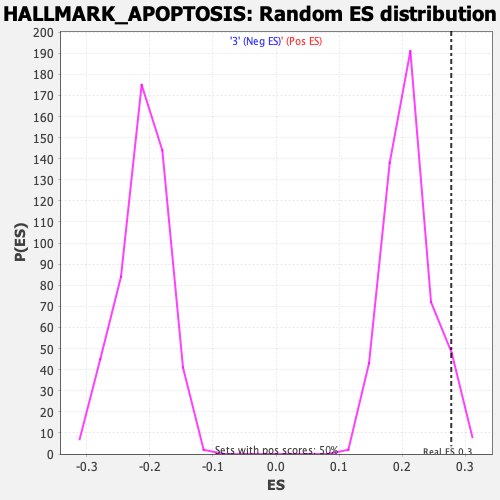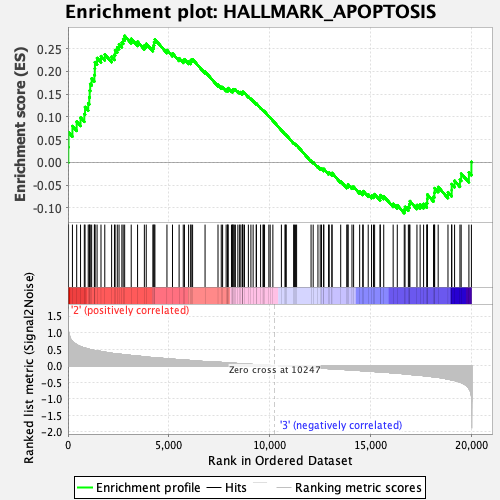
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group3_versus_Group4.GMP.neu_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_APOPTOSIS |
| Enrichment Score (ES) | 0.27770993 |
| Normalized Enrichment Score (NES) | 1.3188864 |
| Nominal p-value | 0.045816734 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.748 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tnfrsf12a | 16 | 1.064 | 0.0333 | Yes |
| 2 | Gsn | 27 | 1.038 | 0.0661 | Yes |
| 3 | Ccnd1 | 220 | 0.724 | 0.0796 | Yes |
| 4 | Mmp2 | 431 | 0.635 | 0.0894 | Yes |
| 5 | Fas | 622 | 0.577 | 0.0983 | Yes |
| 6 | Cdc25b | 809 | 0.539 | 0.1062 | Yes |
| 7 | Bcl2l1 | 845 | 0.531 | 0.1215 | Yes |
| 8 | Eno2 | 1003 | 0.502 | 0.1297 | Yes |
| 9 | Rhob | 1055 | 0.494 | 0.1430 | Yes |
| 10 | Mgmt | 1076 | 0.492 | 0.1577 | Yes |
| 11 | Sc5d | 1101 | 0.488 | 0.1722 | Yes |
| 12 | Gch1 | 1173 | 0.482 | 0.1841 | Yes |
| 13 | Smad7 | 1307 | 0.460 | 0.1921 | Yes |
| 14 | Timp2 | 1330 | 0.457 | 0.2057 | Yes |
| 15 | Bgn | 1337 | 0.457 | 0.2200 | Yes |
| 16 | Gadd45b | 1442 | 0.447 | 0.2291 | Yes |
| 17 | Gpx3 | 1636 | 0.430 | 0.2332 | Yes |
| 18 | Pmaip1 | 1824 | 0.410 | 0.2369 | Yes |
| 19 | Lgals3 | 2163 | 0.374 | 0.2319 | Yes |
| 20 | Tnf | 2304 | 0.365 | 0.2365 | Yes |
| 21 | Cth | 2336 | 0.361 | 0.2465 | Yes |
| 22 | Timp3 | 2444 | 0.356 | 0.2526 | Yes |
| 23 | Ccnd2 | 2531 | 0.349 | 0.2594 | Yes |
| 24 | Txnip | 2672 | 0.339 | 0.2632 | Yes |
| 25 | Irf1 | 2736 | 0.336 | 0.2708 | Yes |
| 26 | Tgfb2 | 2810 | 0.329 | 0.2777 | Yes |
| 27 | Ptk2 | 3135 | 0.309 | 0.2713 | No |
| 28 | Btg2 | 3450 | 0.292 | 0.2649 | No |
| 29 | Pak1 | 3787 | 0.268 | 0.2566 | No |
| 30 | Bid | 3880 | 0.261 | 0.2603 | No |
| 31 | Gpx1 | 4209 | 0.244 | 0.2517 | No |
| 32 | F2r | 4254 | 0.242 | 0.2572 | No |
| 33 | Ifngr1 | 4270 | 0.241 | 0.2642 | No |
| 34 | Sod2 | 4310 | 0.238 | 0.2698 | No |
| 35 | Il18 | 4907 | 0.210 | 0.2466 | No |
| 36 | Sptan1 | 5182 | 0.196 | 0.2391 | No |
| 37 | Pdgfrb | 5510 | 0.184 | 0.2285 | No |
| 38 | Gsr | 5715 | 0.173 | 0.2238 | No |
| 39 | App | 5775 | 0.170 | 0.2263 | No |
| 40 | Mcl1 | 5981 | 0.162 | 0.2212 | No |
| 41 | Casp9 | 6087 | 0.156 | 0.2209 | No |
| 42 | Pdcd4 | 6104 | 0.156 | 0.2251 | No |
| 43 | Add1 | 6179 | 0.152 | 0.2262 | No |
| 44 | Etf1 | 6797 | 0.124 | 0.1992 | No |
| 45 | Gna15 | 7439 | 0.105 | 0.1703 | No |
| 46 | Casp6 | 7600 | 0.099 | 0.1655 | No |
| 47 | Isg20 | 7667 | 0.097 | 0.1652 | No |
| 48 | Tap1 | 7843 | 0.089 | 0.1593 | No |
| 49 | Casp4 | 7921 | 0.086 | 0.1582 | No |
| 50 | Lmna | 7922 | 0.086 | 0.1610 | No |
| 51 | Cd69 | 7938 | 0.086 | 0.1630 | No |
| 52 | Casp7 | 8107 | 0.079 | 0.1571 | No |
| 53 | Cav1 | 8171 | 0.077 | 0.1564 | No |
| 54 | Btg3 | 8174 | 0.077 | 0.1587 | No |
| 55 | Cflar | 8188 | 0.076 | 0.1605 | No |
| 56 | Anxa1 | 8247 | 0.074 | 0.1599 | No |
| 57 | Plcb2 | 8300 | 0.072 | 0.1596 | No |
| 58 | Psen1 | 8424 | 0.067 | 0.1556 | No |
| 59 | Nedd9 | 8518 | 0.063 | 0.1529 | No |
| 60 | Ank | 8533 | 0.062 | 0.1542 | No |
| 61 | Psen2 | 8632 | 0.058 | 0.1511 | No |
| 62 | Top2a | 8660 | 0.057 | 0.1516 | No |
| 63 | Madd | 8667 | 0.057 | 0.1531 | No |
| 64 | Sqstm1 | 8668 | 0.057 | 0.1549 | No |
| 65 | Birc3 | 8751 | 0.053 | 0.1525 | No |
| 66 | Jun | 8945 | 0.046 | 0.1443 | No |
| 67 | Igf2r | 9071 | 0.041 | 0.1393 | No |
| 68 | Vdac2 | 9181 | 0.036 | 0.1350 | No |
| 69 | Fdxr | 9339 | 0.032 | 0.1281 | No |
| 70 | Rock1 | 9340 | 0.032 | 0.1291 | No |
| 71 | H1f0 | 9550 | 0.025 | 0.1194 | No |
| 72 | Casp2 | 9670 | 0.020 | 0.1141 | No |
| 73 | Tgfbr3 | 9700 | 0.019 | 0.1132 | No |
| 74 | Dffa | 9747 | 0.018 | 0.1115 | No |
| 75 | Hmgb2 | 9961 | 0.010 | 0.1011 | No |
| 76 | Cd14 | 10026 | 0.008 | 0.0982 | No |
| 77 | Bcl2l11 | 10160 | 0.003 | 0.0916 | No |
| 78 | Satb1 | 10583 | -0.005 | 0.0705 | No |
| 79 | Bcl2l2 | 10760 | -0.011 | 0.0620 | No |
| 80 | Dap | 10792 | -0.012 | 0.0608 | No |
| 81 | Bmf | 10823 | -0.013 | 0.0597 | No |
| 82 | Casp8 | 11190 | -0.025 | 0.0421 | No |
| 83 | Nefh | 11229 | -0.027 | 0.0411 | No |
| 84 | Retsat | 11286 | -0.029 | 0.0392 | No |
| 85 | Xiap | 11337 | -0.031 | 0.0377 | No |
| 86 | Bax | 12054 | -0.054 | 0.0034 | No |
| 87 | Diablo | 12159 | -0.058 | 0.0000 | No |
| 88 | Ebp | 12395 | -0.066 | -0.0096 | No |
| 89 | Ppp2r5b | 12508 | -0.071 | -0.0130 | No |
| 90 | Bnip3l | 12573 | -0.073 | -0.0139 | No |
| 91 | Ppp3r1 | 12680 | -0.077 | -0.0168 | No |
| 92 | Rnasel | 12684 | -0.077 | -0.0145 | No |
| 93 | Wee1 | 12923 | -0.086 | -0.0237 | No |
| 94 | Dnm1l | 12954 | -0.087 | -0.0224 | No |
| 95 | Erbb3 | 13090 | -0.091 | -0.0263 | No |
| 96 | Cyld | 13092 | -0.091 | -0.0234 | No |
| 97 | Cd44 | 13524 | -0.107 | -0.0417 | No |
| 98 | Dcn | 13823 | -0.118 | -0.0529 | No |
| 99 | Sod1 | 13877 | -0.120 | -0.0517 | No |
| 100 | Crebbp | 13890 | -0.120 | -0.0484 | No |
| 101 | Pea15a | 14083 | -0.129 | -0.0540 | No |
| 102 | Rara | 14164 | -0.132 | -0.0537 | No |
| 103 | Ppt1 | 14464 | -0.144 | -0.0642 | No |
| 104 | Dpyd | 14616 | -0.150 | -0.0670 | No |
| 105 | Ddit3 | 14640 | -0.151 | -0.0633 | No |
| 106 | Krt18 | 14888 | -0.162 | -0.0705 | No |
| 107 | Rela | 15053 | -0.169 | -0.0734 | No |
| 108 | Brca1 | 15149 | -0.172 | -0.0726 | No |
| 109 | Ier3 | 15206 | -0.175 | -0.0699 | No |
| 110 | Cdk2 | 15478 | -0.183 | -0.0776 | No |
| 111 | Dnaja1 | 15485 | -0.184 | -0.0720 | No |
| 112 | Bcl10 | 15658 | -0.191 | -0.0745 | No |
| 113 | Casp3 | 16125 | -0.212 | -0.0912 | No |
| 114 | Ctnnb1 | 16330 | -0.223 | -0.0943 | No |
| 115 | Gadd45a | 16675 | -0.243 | -0.1038 | No |
| 116 | Dap3 | 16714 | -0.245 | -0.0979 | No |
| 117 | Gpx4 | 16880 | -0.255 | -0.0980 | No |
| 118 | Cd38 | 16928 | -0.258 | -0.0921 | No |
| 119 | Casp1 | 16957 | -0.260 | -0.0852 | No |
| 120 | Tnfsf10 | 17305 | -0.277 | -0.0937 | No |
| 121 | Cdkn1b | 17463 | -0.287 | -0.0924 | No |
| 122 | Gstm2 | 17635 | -0.299 | -0.0914 | No |
| 123 | Sat1 | 17793 | -0.310 | -0.0894 | No |
| 124 | Dnajc3 | 17816 | -0.312 | -0.0805 | No |
| 125 | Aifm3 | 17821 | -0.312 | -0.0707 | No |
| 126 | Clu | 18128 | -0.333 | -0.0754 | No |
| 127 | Hmox1 | 18169 | -0.335 | -0.0667 | No |
| 128 | Bcap31 | 18184 | -0.335 | -0.0567 | No |
| 129 | Slc20a1 | 18358 | -0.349 | -0.0542 | No |
| 130 | Lef1 | 18844 | -0.404 | -0.0657 | No |
| 131 | Atf3 | 19027 | -0.424 | -0.0612 | No |
| 132 | Rhot2 | 19030 | -0.424 | -0.0477 | No |
| 133 | Bik | 19172 | -0.444 | -0.0406 | No |
| 134 | Tspo | 19438 | -0.495 | -0.0381 | No |
| 135 | Cdkn1a | 19498 | -0.508 | -0.0248 | No |
| 136 | Hgf | 19882 | -0.685 | -0.0221 | No |
| 137 | Ifitm3 | 20006 | -0.913 | 0.0010 | No |