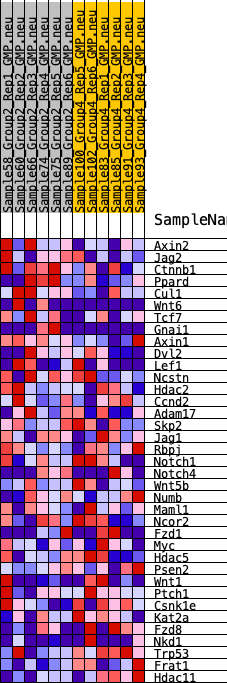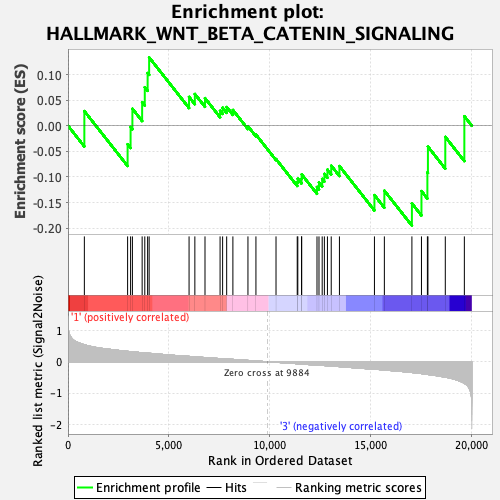
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group2_versus_Group4.GMP.neu_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_WNT_BETA_CATENIN_SIGNALING |
| Enrichment Score (ES) | -0.19515243 |
| Normalized Enrichment Score (NES) | -0.8081177 |
| Nominal p-value | 0.76031435 |
| FDR q-value | 0.90143335 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Axin2 | 811 | 0.544 | 0.0286 | No |
| 2 | Jag2 | 2956 | 0.334 | -0.0362 | No |
| 3 | Ctnnb1 | 3104 | 0.323 | -0.0026 | No |
| 4 | Ppard | 3194 | 0.317 | 0.0332 | No |
| 5 | Cul1 | 3677 | 0.290 | 0.0460 | No |
| 6 | Wnt6 | 3808 | 0.281 | 0.0752 | No |
| 7 | Tcf7 | 3949 | 0.274 | 0.1029 | No |
| 8 | Gnai1 | 4024 | 0.270 | 0.1335 | No |
| 9 | Axin1 | 6004 | 0.172 | 0.0563 | No |
| 10 | Dvl2 | 6291 | 0.157 | 0.0620 | No |
| 11 | Lef1 | 6794 | 0.134 | 0.0539 | No |
| 12 | Ncstn | 7542 | 0.101 | 0.0293 | No |
| 13 | Hdac2 | 7668 | 0.095 | 0.0351 | No |
| 14 | Ccnd2 | 7869 | 0.089 | 0.0365 | No |
| 15 | Adam17 | 8177 | 0.076 | 0.0308 | No |
| 16 | Skp2 | 8924 | 0.044 | -0.0010 | No |
| 17 | Jag1 | 9320 | 0.027 | -0.0174 | No |
| 18 | Rbpj | 10316 | -0.015 | -0.0652 | No |
| 19 | Notch1 | 11368 | -0.060 | -0.1101 | No |
| 20 | Notch4 | 11397 | -0.062 | -0.1036 | No |
| 21 | Wnt5b | 11582 | -0.071 | -0.1039 | No |
| 22 | Numb | 11589 | -0.071 | -0.0952 | No |
| 23 | Maml1 | 12348 | -0.106 | -0.1196 | No |
| 24 | Ncor2 | 12447 | -0.110 | -0.1106 | No |
| 25 | Fzd1 | 12606 | -0.115 | -0.1039 | No |
| 26 | Myc | 12714 | -0.119 | -0.0941 | No |
| 27 | Hdac5 | 12866 | -0.125 | -0.0858 | No |
| 28 | Psen2 | 13054 | -0.135 | -0.0780 | No |
| 29 | Wnt1 | 13461 | -0.153 | -0.0789 | No |
| 30 | Ptch1 | 15197 | -0.237 | -0.1356 | No |
| 31 | Csnk1e | 15689 | -0.262 | -0.1269 | No |
| 32 | Kat2a | 17055 | -0.341 | -0.1518 | Yes |
| 33 | Fzd8 | 17529 | -0.376 | -0.1278 | Yes |
| 34 | Nkd1 | 17823 | -0.403 | -0.0912 | Yes |
| 35 | Trp53 | 17850 | -0.406 | -0.0409 | Yes |
| 36 | Frat1 | 18711 | -0.489 | -0.0218 | Yes |
| 37 | Hdac11 | 19656 | -0.689 | 0.0185 | Yes |