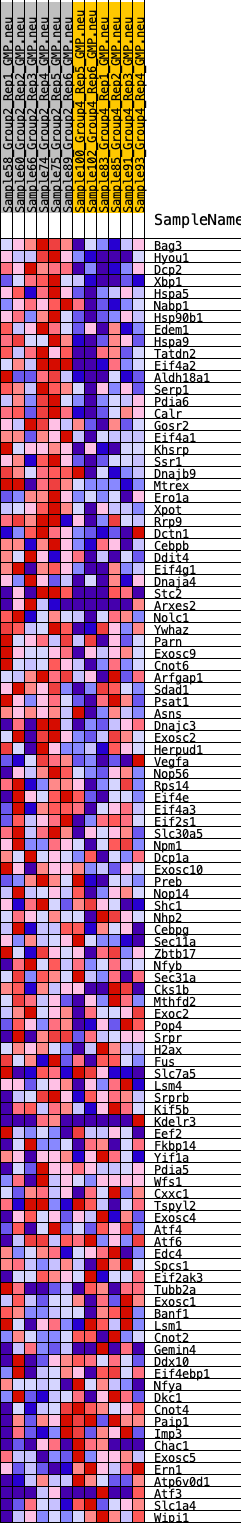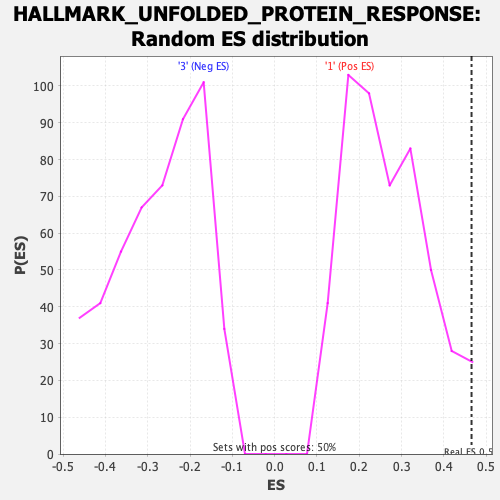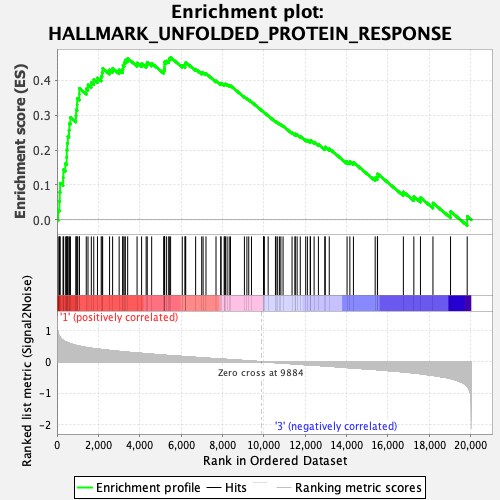
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group2_versus_Group4.GMP.neu_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.46552667 |
| Normalized Enrichment Score (NES) | 1.7595152 |
| Nominal p-value | 0.025948104 |
| FDR q-value | 0.052320592 |
| FWER p-Value | 0.074 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Bag3 | 55 | 0.900 | 0.0279 | Yes |
| 2 | Hyou1 | 115 | 0.819 | 0.0529 | Yes |
| 3 | Dcp2 | 131 | 0.804 | 0.0795 | Yes |
| 4 | Xbp1 | 152 | 0.781 | 0.1052 | Yes |
| 5 | Hspa5 | 296 | 0.683 | 0.1213 | Yes |
| 6 | Nabp1 | 308 | 0.678 | 0.1438 | Yes |
| 7 | Hsp90b1 | 402 | 0.642 | 0.1611 | Yes |
| 8 | Edem1 | 467 | 0.623 | 0.1791 | Yes |
| 9 | Hspa9 | 473 | 0.622 | 0.2001 | Yes |
| 10 | Tatdn2 | 494 | 0.617 | 0.2201 | Yes |
| 11 | Eif4a2 | 523 | 0.611 | 0.2395 | Yes |
| 12 | Aldh18a1 | 580 | 0.597 | 0.2571 | Yes |
| 13 | Serp1 | 598 | 0.591 | 0.2763 | Yes |
| 14 | Pdia6 | 650 | 0.576 | 0.2934 | Yes |
| 15 | Calr | 915 | 0.525 | 0.2981 | Yes |
| 16 | Gosr2 | 926 | 0.523 | 0.3154 | Yes |
| 17 | Eif4a1 | 974 | 0.515 | 0.3306 | Yes |
| 18 | Khsrp | 978 | 0.515 | 0.3480 | Yes |
| 19 | Ssr1 | 1077 | 0.499 | 0.3601 | Yes |
| 20 | Dnajb9 | 1080 | 0.498 | 0.3770 | Yes |
| 21 | Mtrex | 1414 | 0.455 | 0.3758 | Yes |
| 22 | Ero1a | 1498 | 0.447 | 0.3868 | Yes |
| 23 | Xpot | 1659 | 0.431 | 0.3935 | Yes |
| 24 | Rrp9 | 1777 | 0.421 | 0.4020 | Yes |
| 25 | Dctn1 | 1956 | 0.405 | 0.4068 | Yes |
| 26 | Cebpb | 2137 | 0.392 | 0.4112 | Yes |
| 27 | Ddit4 | 2179 | 0.389 | 0.4224 | Yes |
| 28 | Eif4g1 | 2207 | 0.387 | 0.4342 | Yes |
| 29 | Dnaja4 | 2541 | 0.361 | 0.4298 | Yes |
| 30 | Stc2 | 2688 | 0.350 | 0.4344 | Yes |
| 31 | Arxes2 | 3004 | 0.330 | 0.4298 | Yes |
| 32 | Nolc1 | 3174 | 0.318 | 0.4322 | Yes |
| 33 | Ywhaz | 3191 | 0.317 | 0.4422 | Yes |
| 34 | Parn | 3246 | 0.315 | 0.4502 | Yes |
| 35 | Exosc9 | 3304 | 0.312 | 0.4580 | Yes |
| 36 | Cnot6 | 3416 | 0.305 | 0.4628 | Yes |
| 37 | Arfgap1 | 3870 | 0.279 | 0.4496 | Yes |
| 38 | Sdad1 | 4089 | 0.267 | 0.4478 | Yes |
| 39 | Psat1 | 4309 | 0.254 | 0.4454 | Yes |
| 40 | Asns | 4362 | 0.251 | 0.4514 | Yes |
| 41 | Dnajc3 | 4576 | 0.240 | 0.4489 | Yes |
| 42 | Exosc2 | 5158 | 0.210 | 0.4268 | Yes |
| 43 | Herpud1 | 5186 | 0.208 | 0.4326 | Yes |
| 44 | Vegfa | 5197 | 0.207 | 0.4391 | Yes |
| 45 | Nop56 | 5199 | 0.207 | 0.4461 | Yes |
| 46 | Rps14 | 5203 | 0.206 | 0.4530 | Yes |
| 47 | Eif4e | 5299 | 0.203 | 0.4552 | Yes |
| 48 | Eif4a3 | 5402 | 0.196 | 0.4567 | Yes |
| 49 | Eif2s1 | 5428 | 0.195 | 0.4621 | Yes |
| 50 | Slc30a5 | 5491 | 0.191 | 0.4655 | Yes |
| 51 | Npm1 | 6055 | 0.169 | 0.4430 | No |
| 52 | Dcp1a | 6191 | 0.163 | 0.4418 | No |
| 53 | Exosc10 | 6195 | 0.162 | 0.4472 | No |
| 54 | Preb | 6222 | 0.161 | 0.4514 | No |
| 55 | Nop14 | 6701 | 0.139 | 0.4321 | No |
| 56 | Shc1 | 6995 | 0.123 | 0.4216 | No |
| 57 | Nhp2 | 7062 | 0.120 | 0.4224 | No |
| 58 | Cebpg | 7201 | 0.114 | 0.4193 | No |
| 59 | Sec11a | 7686 | 0.094 | 0.3983 | No |
| 60 | Zbtb17 | 7914 | 0.088 | 0.3898 | No |
| 61 | Nfyb | 7931 | 0.087 | 0.3920 | No |
| 62 | Sec31a | 8080 | 0.080 | 0.3873 | No |
| 63 | Cks1b | 8109 | 0.078 | 0.3886 | No |
| 64 | Mthfd2 | 8140 | 0.077 | 0.3897 | No |
| 65 | Exoc2 | 8223 | 0.074 | 0.3881 | No |
| 66 | Pop4 | 8324 | 0.068 | 0.3854 | No |
| 67 | Srpr | 8386 | 0.066 | 0.3846 | No |
| 68 | H2ax | 9059 | 0.038 | 0.3521 | No |
| 69 | Fus | 9179 | 0.032 | 0.3473 | No |
| 70 | Slc7a5 | 9269 | 0.029 | 0.3438 | No |
| 71 | Lsm4 | 9401 | 0.023 | 0.3380 | No |
| 72 | Srprb | 9982 | -0.001 | 0.3089 | No |
| 73 | Kif5b | 9984 | -0.001 | 0.3089 | No |
| 74 | Kdelr3 | 10024 | -0.003 | 0.3071 | No |
| 75 | Eef2 | 10207 | -0.011 | 0.2983 | No |
| 76 | Fkbp14 | 10559 | -0.025 | 0.2815 | No |
| 77 | Yif1a | 10588 | -0.026 | 0.2810 | No |
| 78 | Pdia5 | 10667 | -0.030 | 0.2781 | No |
| 79 | Wfs1 | 10761 | -0.033 | 0.2746 | No |
| 80 | Cxxc1 | 10822 | -0.035 | 0.2728 | No |
| 81 | Tspyl2 | 10928 | -0.041 | 0.2689 | No |
| 82 | Exosc4 | 11362 | -0.060 | 0.2492 | No |
| 83 | Atf4 | 11506 | -0.067 | 0.2443 | No |
| 84 | Atf6 | 11513 | -0.067 | 0.2463 | No |
| 85 | Edc4 | 11607 | -0.072 | 0.2441 | No |
| 86 | Spcs1 | 11759 | -0.078 | 0.2391 | No |
| 87 | Eif2ak3 | 12021 | -0.091 | 0.2292 | No |
| 88 | Tubb2a | 12103 | -0.095 | 0.2283 | No |
| 89 | Exosc1 | 12240 | -0.101 | 0.2250 | No |
| 90 | Banf1 | 12252 | -0.102 | 0.2279 | No |
| 91 | Lsm1 | 12428 | -0.109 | 0.2228 | No |
| 92 | Cnot2 | 12636 | -0.116 | 0.2164 | No |
| 93 | Gemin4 | 12945 | -0.130 | 0.2053 | No |
| 94 | Ddx10 | 12966 | -0.131 | 0.2088 | No |
| 95 | Eif4ebp1 | 13160 | -0.140 | 0.2039 | No |
| 96 | Nfya | 14021 | -0.182 | 0.1669 | No |
| 97 | Dkc1 | 14157 | -0.189 | 0.1666 | No |
| 98 | Cnot4 | 14329 | -0.196 | 0.1647 | No |
| 99 | Paip1 | 15380 | -0.245 | 0.1203 | No |
| 100 | Imp3 | 15488 | -0.250 | 0.1235 | No |
| 101 | Chac1 | 15495 | -0.251 | 0.1317 | No |
| 102 | Exosc5 | 16743 | -0.321 | 0.0801 | No |
| 103 | Ern1 | 17249 | -0.355 | 0.0668 | No |
| 104 | Atp6v0d1 | 17569 | -0.380 | 0.0638 | No |
| 105 | Atf3 | 18173 | -0.432 | 0.0482 | No |
| 106 | Slc1a4 | 19023 | -0.533 | 0.0238 | No |
| 107 | Wipi1 | 19828 | -0.778 | 0.0099 | No |