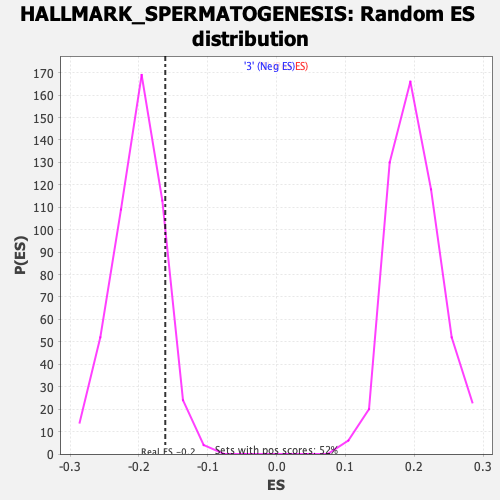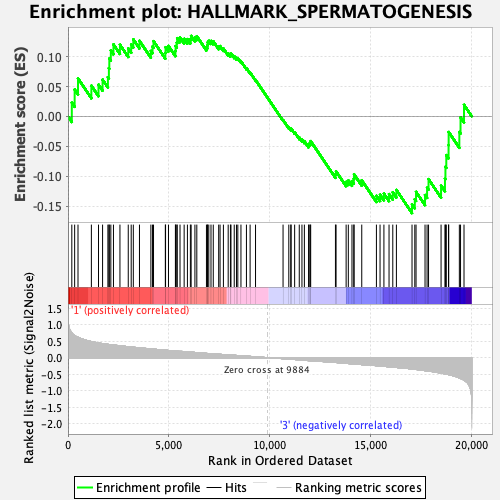
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group2_versus_Group4.GMP.neu_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | -0.161759 |
| Normalized Enrichment Score (NES) | -0.8025078 |
| Nominal p-value | 0.88247424 |
| FDR q-value | 0.85183996 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nek2 | 189 | 0.747 | 0.0230 | No |
| 2 | Dcc | 330 | 0.668 | 0.0449 | No |
| 3 | Sirt1 | 496 | 0.617 | 0.0635 | No |
| 4 | Pgs1 | 1159 | 0.488 | 0.0514 | No |
| 5 | Ybx2 | 1513 | 0.445 | 0.0530 | No |
| 6 | Tsn | 1715 | 0.425 | 0.0614 | No |
| 7 | Gfi1 | 1979 | 0.402 | 0.0657 | No |
| 8 | Scg5 | 2028 | 0.399 | 0.0806 | No |
| 9 | Slc2a5 | 2049 | 0.397 | 0.0969 | No |
| 10 | Bub1 | 2124 | 0.394 | 0.1103 | No |
| 11 | Dbf4 | 2254 | 0.382 | 0.1204 | No |
| 12 | Jam3 | 2576 | 0.358 | 0.1198 | No |
| 13 | Pebp1 | 2988 | 0.331 | 0.1136 | No |
| 14 | Agfg1 | 3134 | 0.321 | 0.1203 | No |
| 15 | Hspa4l | 3239 | 0.315 | 0.1288 | No |
| 16 | Mtor | 3544 | 0.296 | 0.1264 | No |
| 17 | Dmrt1 | 4110 | 0.266 | 0.1096 | No |
| 18 | Stam2 | 4192 | 0.263 | 0.1170 | No |
| 19 | Clvs1 | 4241 | 0.259 | 0.1258 | No |
| 20 | Csnk2a2 | 4827 | 0.227 | 0.1063 | No |
| 21 | Pacrg | 4838 | 0.226 | 0.1156 | No |
| 22 | Pomc | 4983 | 0.218 | 0.1178 | No |
| 23 | Coil | 5326 | 0.201 | 0.1094 | No |
| 24 | Rfc4 | 5336 | 0.200 | 0.1176 | No |
| 25 | Adam2 | 5395 | 0.196 | 0.1233 | No |
| 26 | Parp2 | 5416 | 0.196 | 0.1308 | No |
| 27 | Scg3 | 5555 | 0.189 | 0.1320 | No |
| 28 | Ttk | 5760 | 0.182 | 0.1297 | No |
| 29 | Hspa2 | 5922 | 0.174 | 0.1292 | No |
| 30 | Topbp1 | 6069 | 0.168 | 0.1292 | No |
| 31 | Ddx4 | 6106 | 0.167 | 0.1346 | No |
| 32 | Nos1 | 6294 | 0.157 | 0.1321 | No |
| 33 | Cdk1 | 6388 | 0.153 | 0.1340 | No |
| 34 | Aurka | 6866 | 0.129 | 0.1157 | No |
| 35 | Zc3h14 | 6913 | 0.127 | 0.1189 | No |
| 36 | Cnih2 | 6926 | 0.126 | 0.1238 | No |
| 37 | Ift88 | 6982 | 0.124 | 0.1264 | No |
| 38 | Nefh | 7091 | 0.119 | 0.1262 | No |
| 39 | Septin4 | 7204 | 0.114 | 0.1256 | No |
| 40 | Oaz3 | 7473 | 0.104 | 0.1166 | No |
| 41 | Ccnb2 | 7541 | 0.101 | 0.1176 | No |
| 42 | Ldhc | 7707 | 0.094 | 0.1134 | No |
| 43 | Art3 | 7944 | 0.086 | 0.1053 | No |
| 44 | Ide | 8055 | 0.081 | 0.1033 | No |
| 45 | Kif2c | 8082 | 0.080 | 0.1055 | No |
| 46 | Ncaph | 8244 | 0.073 | 0.1006 | No |
| 47 | Hspa1l | 8355 | 0.067 | 0.0980 | No |
| 48 | Vdac3 | 8422 | 0.064 | 0.0974 | No |
| 49 | Mast2 | 8577 | 0.059 | 0.0922 | No |
| 50 | Prkar2a | 8849 | 0.047 | 0.0807 | No |
| 51 | Cdkn3 | 9031 | 0.039 | 0.0733 | No |
| 52 | Tle4 | 9300 | 0.028 | 0.0610 | No |
| 53 | Gmcl1 | 10665 | -0.030 | -0.0061 | No |
| 54 | Mllt10 | 10950 | -0.042 | -0.0185 | No |
| 55 | Ezh2 | 11050 | -0.045 | -0.0215 | No |
| 56 | Chfr | 11063 | -0.046 | -0.0201 | No |
| 57 | Arl4a | 11240 | -0.054 | -0.0266 | No |
| 58 | Cct6b | 11469 | -0.065 | -0.0352 | No |
| 59 | Nf2 | 11602 | -0.071 | -0.0388 | No |
| 60 | Psmg1 | 11722 | -0.076 | -0.0414 | No |
| 61 | Dmc1 | 11931 | -0.088 | -0.0480 | No |
| 62 | Pias2 | 11959 | -0.089 | -0.0455 | No |
| 63 | Rad17 | 12013 | -0.091 | -0.0442 | No |
| 64 | Gapdhs | 12035 | -0.092 | -0.0413 | No |
| 65 | Mlf1 | 13263 | -0.144 | -0.0966 | No |
| 66 | She | 13294 | -0.145 | -0.0918 | No |
| 67 | Tnni3 | 13793 | -0.171 | -0.1094 | No |
| 68 | Strbp | 13902 | -0.176 | -0.1071 | No |
| 69 | Camk4 | 14083 | -0.185 | -0.1081 | No |
| 70 | Pcsk4 | 14174 | -0.189 | -0.1044 | No |
| 71 | Clpb | 14187 | -0.190 | -0.0967 | No |
| 72 | Braf | 14568 | -0.208 | -0.1068 | No |
| 73 | Ip6k1 | 15295 | -0.241 | -0.1327 | No |
| 74 | Map7 | 15475 | -0.250 | -0.1309 | No |
| 75 | Spata6 | 15667 | -0.261 | -0.1291 | No |
| 76 | Tulp2 | 15921 | -0.275 | -0.1299 | No |
| 77 | Cftr | 16107 | -0.285 | -0.1268 | No |
| 78 | Grm8 | 16286 | -0.295 | -0.1229 | No |
| 79 | Phf7 | 17061 | -0.342 | -0.1469 | Yes |
| 80 | Phkg2 | 17199 | -0.351 | -0.1385 | Yes |
| 81 | Gstm5 | 17260 | -0.356 | -0.1261 | Yes |
| 82 | Zpbp | 17706 | -0.392 | -0.1314 | Yes |
| 83 | Nphp1 | 17808 | -0.402 | -0.1190 | Yes |
| 84 | Acrbp | 17880 | -0.408 | -0.1049 | Yes |
| 85 | Zc2hc1c | 18501 | -0.468 | -0.1156 | Yes |
| 86 | Slc12a2 | 18695 | -0.487 | -0.1042 | Yes |
| 87 | Taldo1 | 18718 | -0.489 | -0.0841 | Yes |
| 88 | Crisp2 | 18761 | -0.494 | -0.0647 | Yes |
| 89 | Adad1 | 18873 | -0.510 | -0.0481 | Yes |
| 90 | Il12rb2 | 18878 | -0.512 | -0.0261 | Yes |
| 91 | Lpin1 | 19408 | -0.613 | -0.0260 | Yes |
| 92 | Tcp11 | 19467 | -0.628 | -0.0016 | Yes |
| 93 | Tekt2 | 19639 | -0.682 | 0.0194 | Yes |