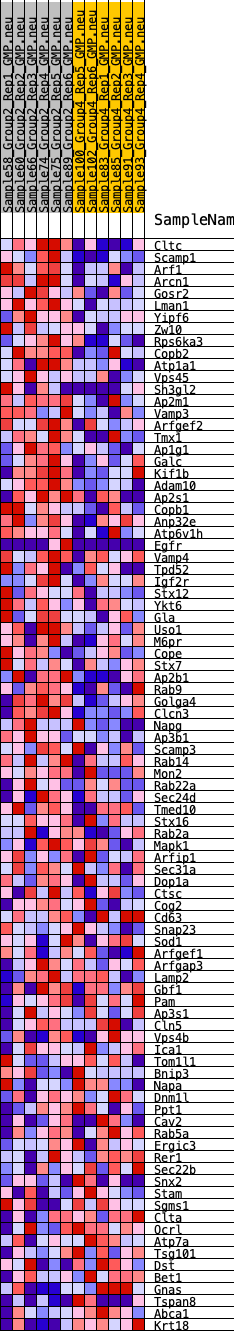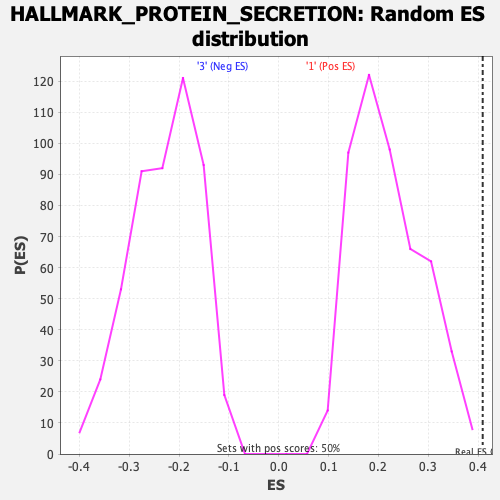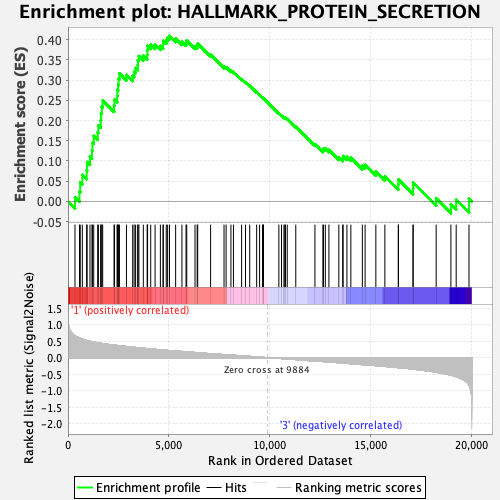
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group2_versus_Group4.GMP.neu_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.4092946 |
| Normalized Enrichment Score (NES) | 1.8539345 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.027318452 |
| FWER p-Value | 0.029 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cltc | 346 | 0.662 | 0.0104 | Yes |
| 2 | Scamp1 | 570 | 0.599 | 0.0242 | Yes |
| 3 | Arf1 | 607 | 0.588 | 0.0470 | Yes |
| 4 | Arcn1 | 710 | 0.564 | 0.0655 | Yes |
| 5 | Gosr2 | 926 | 0.523 | 0.0766 | Yes |
| 6 | Lman1 | 946 | 0.519 | 0.0974 | Yes |
| 7 | Yipf6 | 1090 | 0.497 | 0.1110 | Yes |
| 8 | Zw10 | 1190 | 0.483 | 0.1263 | Yes |
| 9 | Rps6ka3 | 1212 | 0.480 | 0.1453 | Yes |
| 10 | Copb2 | 1276 | 0.471 | 0.1619 | Yes |
| 11 | Atp1a1 | 1472 | 0.450 | 0.1709 | Yes |
| 12 | Vps45 | 1507 | 0.446 | 0.1879 | Yes |
| 13 | Sh3gl2 | 1621 | 0.434 | 0.2004 | Yes |
| 14 | Ap2m1 | 1636 | 0.432 | 0.2178 | Yes |
| 15 | Vamp3 | 1677 | 0.430 | 0.2338 | Yes |
| 16 | Arfgef2 | 1720 | 0.425 | 0.2494 | Yes |
| 17 | Tmx1 | 2283 | 0.380 | 0.2372 | Yes |
| 18 | Ap1g1 | 2307 | 0.378 | 0.2518 | Yes |
| 19 | Galc | 2436 | 0.370 | 0.2609 | Yes |
| 20 | Kif1b | 2449 | 0.369 | 0.2757 | Yes |
| 21 | Adam10 | 2490 | 0.366 | 0.2890 | Yes |
| 22 | Ap2s1 | 2508 | 0.364 | 0.3034 | Yes |
| 23 | Copb1 | 2551 | 0.359 | 0.3164 | Yes |
| 24 | Anp32e | 2899 | 0.337 | 0.3130 | Yes |
| 25 | Atp6v1h | 3208 | 0.316 | 0.3108 | Yes |
| 26 | Egfr | 3279 | 0.313 | 0.3204 | Yes |
| 27 | Vamp4 | 3352 | 0.309 | 0.3297 | Yes |
| 28 | Tpd52 | 3460 | 0.303 | 0.3370 | Yes |
| 29 | Igf2r | 3468 | 0.302 | 0.3493 | Yes |
| 30 | Stx12 | 3516 | 0.298 | 0.3595 | Yes |
| 31 | Ykt6 | 3735 | 0.286 | 0.3605 | Yes |
| 32 | Gla | 3930 | 0.275 | 0.3623 | Yes |
| 33 | Uso1 | 3936 | 0.275 | 0.3735 | Yes |
| 34 | M6pr | 3947 | 0.274 | 0.3845 | Yes |
| 35 | Cope | 4102 | 0.267 | 0.3879 | Yes |
| 36 | Stx7 | 4319 | 0.253 | 0.3877 | Yes |
| 37 | Ap2b1 | 4581 | 0.240 | 0.3846 | Yes |
| 38 | Rab9 | 4717 | 0.233 | 0.3876 | Yes |
| 39 | Golga4 | 4721 | 0.233 | 0.3972 | Yes |
| 40 | Clcn3 | 4873 | 0.224 | 0.3990 | Yes |
| 41 | Napg | 4941 | 0.220 | 0.4048 | Yes |
| 42 | Ap3b1 | 5033 | 0.215 | 0.4093 | Yes |
| 43 | Scamp3 | 5338 | 0.200 | 0.4024 | No |
| 44 | Rab14 | 5651 | 0.189 | 0.3947 | No |
| 45 | Mon2 | 5853 | 0.178 | 0.3920 | No |
| 46 | Rab22a | 5889 | 0.176 | 0.3976 | No |
| 47 | Sec24d | 6299 | 0.157 | 0.3837 | No |
| 48 | Tmed10 | 6406 | 0.152 | 0.3847 | No |
| 49 | Stx16 | 6433 | 0.150 | 0.3897 | No |
| 50 | Rab2a | 7072 | 0.120 | 0.3627 | No |
| 51 | Mapk1 | 7738 | 0.092 | 0.3332 | No |
| 52 | Arfip1 | 7841 | 0.091 | 0.3319 | No |
| 53 | Sec31a | 8080 | 0.080 | 0.3233 | No |
| 54 | Dop1a | 8205 | 0.075 | 0.3202 | No |
| 55 | Ctsc | 8610 | 0.057 | 0.3023 | No |
| 56 | Cog2 | 8803 | 0.049 | 0.2947 | No |
| 57 | Cd63 | 9011 | 0.039 | 0.2860 | No |
| 58 | Snap23 | 9356 | 0.025 | 0.2698 | No |
| 59 | Sod1 | 9501 | 0.018 | 0.2633 | No |
| 60 | Arfgef1 | 9645 | 0.011 | 0.2566 | No |
| 61 | Arfgap3 | 9685 | 0.009 | 0.2550 | No |
| 62 | Lamp2 | 9696 | 0.009 | 0.2549 | No |
| 63 | Gbf1 | 10457 | -0.021 | 0.2176 | No |
| 64 | Pam | 10589 | -0.026 | 0.2122 | No |
| 65 | Ap3s1 | 10709 | -0.031 | 0.2075 | No |
| 66 | Cln5 | 10752 | -0.033 | 0.2067 | No |
| 67 | Vps4b | 10784 | -0.034 | 0.2066 | No |
| 68 | Ica1 | 10872 | -0.038 | 0.2038 | No |
| 69 | Tom1l1 | 11296 | -0.057 | 0.1850 | No |
| 70 | Bnip3 | 12246 | -0.102 | 0.1416 | No |
| 71 | Napa | 12642 | -0.116 | 0.1267 | No |
| 72 | Dnm1l | 12668 | -0.117 | 0.1303 | No |
| 73 | Ppt1 | 12761 | -0.121 | 0.1308 | No |
| 74 | Cav2 | 12939 | -0.130 | 0.1273 | No |
| 75 | Rab5a | 13431 | -0.152 | 0.1091 | No |
| 76 | Ergic3 | 13624 | -0.162 | 0.1062 | No |
| 77 | Rer1 | 13643 | -0.163 | 0.1121 | No |
| 78 | Sec22b | 13833 | -0.173 | 0.1099 | No |
| 79 | Snx2 | 14027 | -0.182 | 0.1079 | No |
| 80 | Stam | 14597 | -0.211 | 0.0881 | No |
| 81 | Sgms1 | 14734 | -0.216 | 0.0904 | No |
| 82 | Clta | 15267 | -0.239 | 0.0737 | No |
| 83 | Ocrl | 15714 | -0.263 | 0.0623 | No |
| 84 | Atp7a | 16381 | -0.301 | 0.0415 | No |
| 85 | Tsg101 | 16386 | -0.302 | 0.0540 | No |
| 86 | Dst | 17111 | -0.345 | 0.0321 | No |
| 87 | Bet1 | 17116 | -0.345 | 0.0463 | No |
| 88 | Gnas | 18258 | -0.441 | 0.0075 | No |
| 89 | Tspan8 | 18989 | -0.529 | -0.0070 | No |
| 90 | Abca1 | 19251 | -0.575 | 0.0040 | No |
| 91 | Krt18 | 19886 | -0.832 | 0.0070 | No |