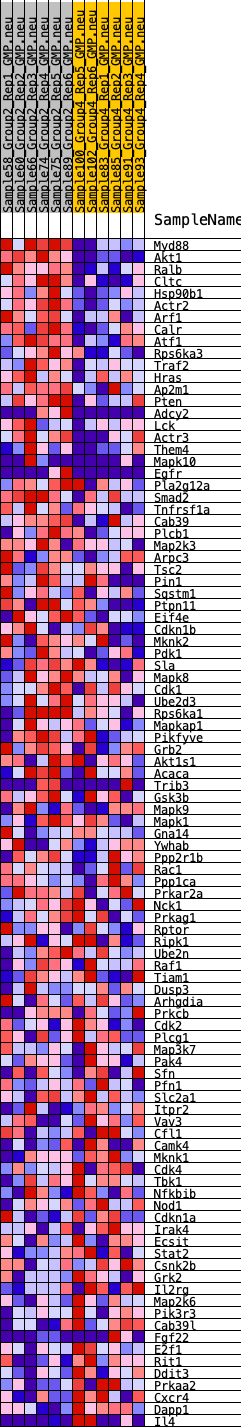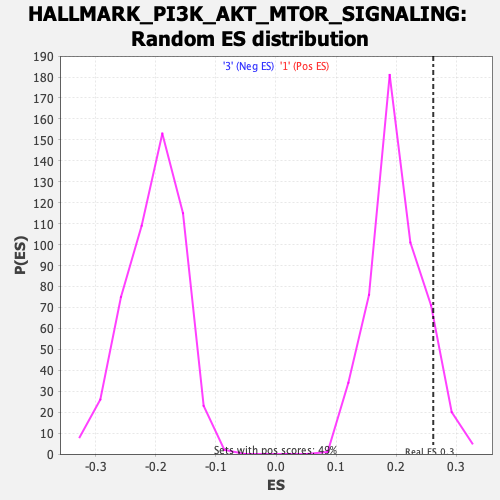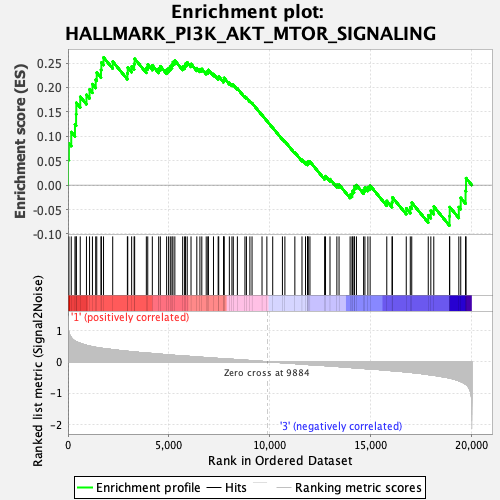
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group2_versus_Group4.GMP.neu_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.26146233 |
| Normalized Enrichment Score (NES) | 1.2957475 |
| Nominal p-value | 0.096114516 |
| FDR q-value | 0.49988237 |
| FWER p-Value | 0.763 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Myd88 | 1 | 1.425 | 0.0531 | Yes |
| 2 | Akt1 | 45 | 0.931 | 0.0856 | Yes |
| 3 | Ralb | 161 | 0.775 | 0.1087 | Yes |
| 4 | Cltc | 346 | 0.662 | 0.1242 | Yes |
| 5 | Hsp90b1 | 402 | 0.642 | 0.1454 | Yes |
| 6 | Actr2 | 412 | 0.640 | 0.1687 | Yes |
| 7 | Arf1 | 607 | 0.588 | 0.1809 | Yes |
| 8 | Calr | 915 | 0.525 | 0.1851 | Yes |
| 9 | Atf1 | 1069 | 0.500 | 0.1960 | Yes |
| 10 | Rps6ka3 | 1212 | 0.480 | 0.2068 | Yes |
| 11 | Traf2 | 1374 | 0.460 | 0.2159 | Yes |
| 12 | Hras | 1426 | 0.454 | 0.2303 | Yes |
| 13 | Ap2m1 | 1636 | 0.432 | 0.2359 | Yes |
| 14 | Pten | 1655 | 0.431 | 0.2511 | Yes |
| 15 | Adcy2 | 1762 | 0.421 | 0.2615 | Yes |
| 16 | Lck | 2217 | 0.385 | 0.2530 | No |
| 17 | Actr3 | 2944 | 0.335 | 0.2291 | No |
| 18 | Them4 | 2963 | 0.333 | 0.2406 | No |
| 19 | Mapk10 | 3156 | 0.319 | 0.2429 | No |
| 20 | Egfr | 3279 | 0.313 | 0.2484 | No |
| 21 | Pla2g12a | 3306 | 0.312 | 0.2587 | No |
| 22 | Smad2 | 3885 | 0.278 | 0.2401 | No |
| 23 | Tnfrsf1a | 3955 | 0.273 | 0.2468 | No |
| 24 | Cab39 | 4182 | 0.263 | 0.2453 | No |
| 25 | Plcb1 | 4497 | 0.242 | 0.2386 | No |
| 26 | Map2k3 | 4579 | 0.240 | 0.2434 | No |
| 27 | Arpc3 | 4891 | 0.223 | 0.2361 | No |
| 28 | Tsc2 | 4989 | 0.218 | 0.2394 | No |
| 29 | Pin1 | 5073 | 0.213 | 0.2432 | No |
| 30 | Sqstm1 | 5156 | 0.210 | 0.2469 | No |
| 31 | Ptpn11 | 5205 | 0.206 | 0.2522 | No |
| 32 | Eif4e | 5299 | 0.203 | 0.2550 | No |
| 33 | Cdkn1b | 5687 | 0.187 | 0.2426 | No |
| 34 | Mknk2 | 5783 | 0.181 | 0.2446 | No |
| 35 | Pdk1 | 5830 | 0.178 | 0.2489 | No |
| 36 | Sla | 5913 | 0.175 | 0.2513 | No |
| 37 | Mapk8 | 6101 | 0.167 | 0.2482 | No |
| 38 | Cdk1 | 6388 | 0.153 | 0.2395 | No |
| 39 | Ube2d3 | 6542 | 0.148 | 0.2373 | No |
| 40 | Rps6ka1 | 6636 | 0.142 | 0.2380 | No |
| 41 | Mapkap1 | 6854 | 0.130 | 0.2319 | No |
| 42 | Pikfyve | 6933 | 0.126 | 0.2327 | No |
| 43 | Grb2 | 6963 | 0.125 | 0.2359 | No |
| 44 | Akt1s1 | 7215 | 0.113 | 0.2275 | No |
| 45 | Acaca | 7447 | 0.105 | 0.2198 | No |
| 46 | Trib3 | 7474 | 0.104 | 0.2224 | No |
| 47 | Gsk3b | 7712 | 0.093 | 0.2140 | No |
| 48 | Mapk9 | 7737 | 0.092 | 0.2162 | No |
| 49 | Mapk1 | 7738 | 0.092 | 0.2196 | No |
| 50 | Gna14 | 8002 | 0.083 | 0.2095 | No |
| 51 | Ywhab | 8130 | 0.078 | 0.2060 | No |
| 52 | Ppp2r1b | 8197 | 0.075 | 0.2055 | No |
| 53 | Rac1 | 8400 | 0.065 | 0.1978 | No |
| 54 | Ppp1ca | 8775 | 0.050 | 0.1809 | No |
| 55 | Prkar2a | 8849 | 0.047 | 0.1790 | No |
| 56 | Nck1 | 9020 | 0.039 | 0.1719 | No |
| 57 | Prkag1 | 9127 | 0.034 | 0.1679 | No |
| 58 | Rptor | 9622 | 0.012 | 0.1435 | No |
| 59 | Ripk1 | 9862 | 0.001 | 0.1316 | No |
| 60 | Ube2n | 10152 | -0.009 | 0.1174 | No |
| 61 | Raf1 | 10634 | -0.029 | 0.0943 | No |
| 62 | Tiam1 | 10756 | -0.033 | 0.0895 | No |
| 63 | Dusp3 | 11250 | -0.055 | 0.0668 | No |
| 64 | Arhgdia | 11604 | -0.071 | 0.0517 | No |
| 65 | Prkcb | 11779 | -0.080 | 0.0460 | No |
| 66 | Cdk2 | 11877 | -0.085 | 0.0443 | No |
| 67 | Plcg1 | 11883 | -0.085 | 0.0472 | No |
| 68 | Map3k7 | 11915 | -0.087 | 0.0489 | No |
| 69 | Pak4 | 11999 | -0.090 | 0.0481 | No |
| 70 | Sfn | 12726 | -0.120 | 0.0161 | No |
| 71 | Pfn1 | 12771 | -0.122 | 0.0184 | No |
| 72 | Slc2a1 | 12993 | -0.132 | 0.0123 | No |
| 73 | Itpr2 | 13334 | -0.147 | 0.0007 | No |
| 74 | Vav3 | 13442 | -0.152 | 0.0010 | No |
| 75 | Cfl1 | 13991 | -0.180 | -0.0198 | No |
| 76 | Camk4 | 14083 | -0.185 | -0.0174 | No |
| 77 | Mknk1 | 14114 | -0.187 | -0.0120 | No |
| 78 | Cdk4 | 14186 | -0.190 | -0.0085 | No |
| 79 | Tbk1 | 14204 | -0.191 | -0.0022 | No |
| 80 | Nfkbib | 14304 | -0.195 | 0.0001 | No |
| 81 | Nod1 | 14660 | -0.213 | -0.0098 | No |
| 82 | Cdkn1a | 14725 | -0.216 | -0.0049 | No |
| 83 | Irak4 | 14872 | -0.223 | -0.0039 | No |
| 84 | Ecsit | 14983 | -0.228 | -0.0009 | No |
| 85 | Stat2 | 15809 | -0.269 | -0.0323 | No |
| 86 | Csnk2b | 16069 | -0.283 | -0.0348 | No |
| 87 | Grk2 | 16089 | -0.284 | -0.0251 | No |
| 88 | Il2rg | 16775 | -0.323 | -0.0475 | No |
| 89 | Map2k6 | 16975 | -0.335 | -0.0450 | No |
| 90 | Pik3r3 | 17047 | -0.341 | -0.0358 | No |
| 91 | Cab39l | 17866 | -0.408 | -0.0617 | No |
| 92 | Fgf22 | 17994 | -0.415 | -0.0526 | No |
| 93 | E2f1 | 18140 | -0.430 | -0.0438 | No |
| 94 | Rit1 | 18919 | -0.517 | -0.0636 | No |
| 95 | Ddit3 | 18932 | -0.518 | -0.0449 | No |
| 96 | Prkaa2 | 19382 | -0.607 | -0.0448 | No |
| 97 | Cxcr4 | 19477 | -0.631 | -0.0260 | No |
| 98 | Dapp1 | 19716 | -0.710 | -0.0115 | No |
| 99 | Il4 | 19740 | -0.724 | 0.0144 | No |