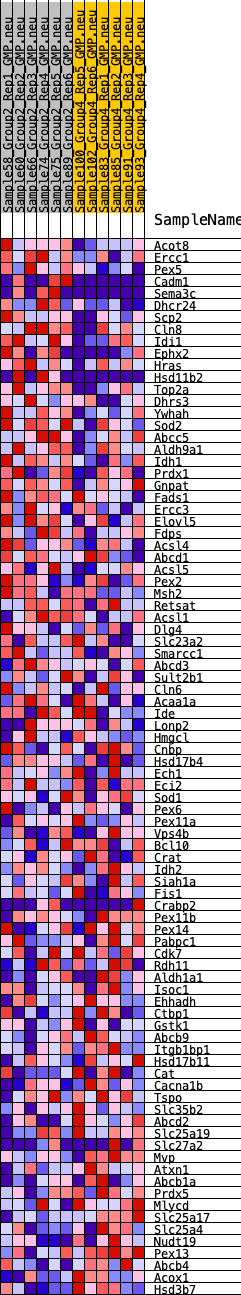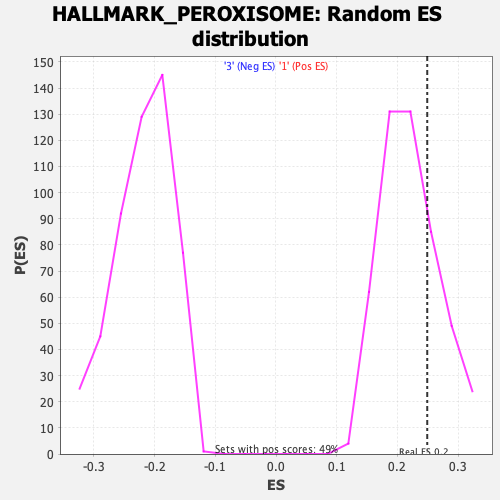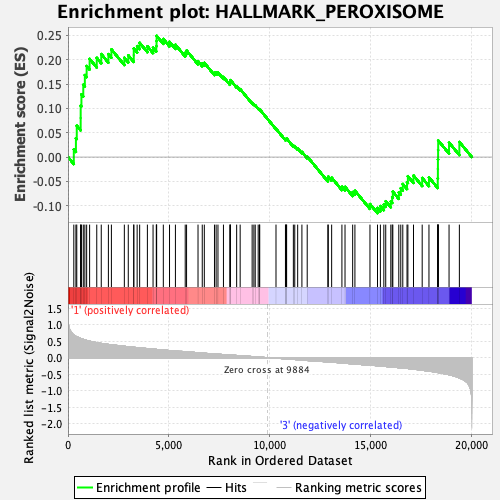
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.neu_Pheno.cls #Group2_versus_Group4.GMP.neu_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | GMP.neu_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | 0.24900903 |
| Normalized Enrichment Score (NES) | 1.1290221 |
| Nominal p-value | 0.28600824 |
| FDR q-value | 0.61829656 |
| FWER p-Value | 0.919 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Acot8 | 288 | 0.686 | 0.0156 | Yes |
| 2 | Ercc1 | 393 | 0.645 | 0.0387 | Yes |
| 3 | Pex5 | 435 | 0.632 | 0.0644 | Yes |
| 4 | Cadm1 | 628 | 0.581 | 0.0802 | Yes |
| 5 | Sema3c | 629 | 0.580 | 0.1056 | Yes |
| 6 | Dhcr24 | 666 | 0.574 | 0.1289 | Yes |
| 7 | Scp2 | 758 | 0.555 | 0.1487 | Yes |
| 8 | Cln8 | 836 | 0.540 | 0.1685 | Yes |
| 9 | Idi1 | 925 | 0.523 | 0.1871 | Yes |
| 10 | Ephx2 | 1072 | 0.500 | 0.2016 | Yes |
| 11 | Hras | 1426 | 0.454 | 0.2039 | Yes |
| 12 | Hsd11b2 | 1649 | 0.431 | 0.2116 | Yes |
| 13 | Top2a | 2003 | 0.401 | 0.2115 | Yes |
| 14 | Dhrs3 | 2152 | 0.391 | 0.2212 | Yes |
| 15 | Ywhah | 2794 | 0.345 | 0.2042 | Yes |
| 16 | Sod2 | 2989 | 0.331 | 0.2090 | Yes |
| 17 | Abcc5 | 3261 | 0.314 | 0.2091 | Yes |
| 18 | Aldh9a1 | 3262 | 0.314 | 0.2229 | Yes |
| 19 | Idh1 | 3429 | 0.304 | 0.2279 | Yes |
| 20 | Prdx1 | 3554 | 0.296 | 0.2347 | Yes |
| 21 | Gnpat | 3937 | 0.275 | 0.2275 | Yes |
| 22 | Fads1 | 4218 | 0.261 | 0.2249 | Yes |
| 23 | Ercc3 | 4379 | 0.250 | 0.2279 | Yes |
| 24 | Elovl5 | 4390 | 0.250 | 0.2383 | Yes |
| 25 | Fdps | 4396 | 0.250 | 0.2490 | Yes |
| 26 | Acsl4 | 4727 | 0.233 | 0.2427 | No |
| 27 | Abcd1 | 5035 | 0.215 | 0.2367 | No |
| 28 | Acsl5 | 5327 | 0.201 | 0.2309 | No |
| 29 | Pex2 | 5813 | 0.180 | 0.2144 | No |
| 30 | Msh2 | 5880 | 0.177 | 0.2189 | No |
| 31 | Retsat | 6448 | 0.150 | 0.1970 | No |
| 32 | Acsl1 | 6658 | 0.141 | 0.1927 | No |
| 33 | Dlg4 | 6760 | 0.136 | 0.1936 | No |
| 34 | Slc23a2 | 7264 | 0.110 | 0.1732 | No |
| 35 | Smarcc1 | 7345 | 0.110 | 0.1740 | No |
| 36 | Abcd3 | 7437 | 0.105 | 0.1740 | No |
| 37 | Sult2b1 | 7709 | 0.094 | 0.1646 | No |
| 38 | Cln6 | 8031 | 0.082 | 0.1520 | No |
| 39 | Acaa1a | 8041 | 0.081 | 0.1552 | No |
| 40 | Ide | 8055 | 0.081 | 0.1581 | No |
| 41 | Lonp2 | 8364 | 0.067 | 0.1455 | No |
| 42 | Hmgcl | 8538 | 0.060 | 0.1395 | No |
| 43 | Cnbp | 9132 | 0.034 | 0.1113 | No |
| 44 | Hsd17b4 | 9219 | 0.031 | 0.1083 | No |
| 45 | Ech1 | 9285 | 0.028 | 0.1063 | No |
| 46 | Eci2 | 9432 | 0.021 | 0.0999 | No |
| 47 | Sod1 | 9501 | 0.018 | 0.0973 | No |
| 48 | Pex6 | 9505 | 0.018 | 0.0979 | No |
| 49 | Pex11a | 10315 | -0.015 | 0.0580 | No |
| 50 | Vps4b | 10784 | -0.034 | 0.0360 | No |
| 51 | Bcl10 | 10795 | -0.034 | 0.0370 | No |
| 52 | Crat | 10834 | -0.036 | 0.0367 | No |
| 53 | Idh2 | 10842 | -0.036 | 0.0379 | No |
| 54 | Siah1a | 11176 | -0.051 | 0.0234 | No |
| 55 | Fis1 | 11245 | -0.054 | 0.0224 | No |
| 56 | Crabp2 | 11390 | -0.062 | 0.0179 | No |
| 57 | Pex11b | 11600 | -0.071 | 0.0105 | No |
| 58 | Pex14 | 11863 | -0.084 | 0.0011 | No |
| 59 | Pabpc1 | 12887 | -0.126 | -0.0447 | No |
| 60 | Cdk7 | 12908 | -0.128 | -0.0401 | No |
| 61 | Rdh11 | 13075 | -0.136 | -0.0425 | No |
| 62 | Aldh1a1 | 13584 | -0.160 | -0.0609 | No |
| 63 | Isoc1 | 13738 | -0.168 | -0.0613 | No |
| 64 | Ehhadh | 14117 | -0.187 | -0.0720 | No |
| 65 | Ctbp1 | 14229 | -0.192 | -0.0692 | No |
| 66 | Gstk1 | 14975 | -0.228 | -0.0966 | No |
| 67 | Abcb9 | 15349 | -0.244 | -0.1046 | No |
| 68 | Itgb1bp1 | 15492 | -0.250 | -0.1007 | No |
| 69 | Hsd17b11 | 15663 | -0.261 | -0.0978 | No |
| 70 | Cat | 15757 | -0.266 | -0.0908 | No |
| 71 | Cacna1b | 16009 | -0.280 | -0.0912 | No |
| 72 | Tspo | 16078 | -0.283 | -0.0822 | No |
| 73 | Slc35b2 | 16109 | -0.285 | -0.0712 | No |
| 74 | Abcd2 | 16405 | -0.303 | -0.0727 | No |
| 75 | Slc25a19 | 16503 | -0.307 | -0.0641 | No |
| 76 | Slc27a2 | 16603 | -0.312 | -0.0554 | No |
| 77 | Mvp | 16813 | -0.325 | -0.0516 | No |
| 78 | Atxn1 | 16856 | -0.328 | -0.0393 | No |
| 79 | Abcb1a | 17134 | -0.346 | -0.0381 | No |
| 80 | Prdx5 | 17565 | -0.379 | -0.0430 | No |
| 81 | Mlycd | 17901 | -0.409 | -0.0419 | No |
| 82 | Slc25a17 | 18337 | -0.449 | -0.0440 | No |
| 83 | Slc25a4 | 18344 | -0.450 | -0.0246 | No |
| 84 | Nudt19 | 18346 | -0.450 | -0.0049 | No |
| 85 | Pex13 | 18355 | -0.450 | 0.0144 | No |
| 86 | Abcb4 | 18359 | -0.451 | 0.0340 | No |
| 87 | Acox1 | 18897 | -0.515 | 0.0297 | No |
| 88 | Hsd3b7 | 19409 | -0.614 | 0.0309 | No |